Background: Klebsiella pneumoniae is considered an opportunistic pathogen associated with nosocomial infections occurring mainly in pediatric patients, such as premature infants placed in intensive care units. The aim of this study was to characterize K. pneumoniae strains isolated from different clinical sources based on their resistance to antibiotics and the presence of virulence factors associated with their persistence in the hospital environment.
Methods: Fifty clinical strains of K. pneumoniae isolated from urine, blood, catheters, and cerebrospinal fluid sources were characterized. Susceptibility testing of antibiotics was performed by the Kirby-Bauer method (Clinical Laboratory Standards Institute, 2010). The ability to form a biofilm was determined by the 96-well microplate method. Capsule and fimbrial structures were visualized by transmission electron microscopy (TEM). Adherence was evaluated on A549 and HT29 cells. Assessment for the presence and expression of the ecpA, fimH, and mrkA genes was performed by PCR and RT-PCR.
Results: Clinical strains of K. pneumoniae were isolated from 48% of urine, 24% of blood, 18% of catheters, and 10% of cerebrospinal fluid. Ninety-two percent of the strains showed resistance to cefpodoxime, whereas few strains showed resistance to imipenem and meropenem (4 and 2%, respectively). The extended spectrum-type beta-lactamase (ESBL) phenotype was identified in 97% of the strains positive for resistance to third-generation cephalosporins. In addition, 88% of the strains were multidrug resistant. All strains were able to form biofilms. Capsule and fimbirial structures were visualized by TEM. Based on our adhesion assays, the A549 cell line was more permissive to K. pneumoniae strains than the HT-29 cell line. K. pneumoniae strains amplified and expressed ecpA (100/70%), fimH (98/2%), and mrkA (84/48%) genes, respectively.
Conclusion: The K. pneumoniae strains exhibited features that allowed them to survive in the hospital environment (formation of biofilm) and resist antimicrobial therapy (multidrug resistant MDR strains). These strains also possessed a capsule, adhesive properties, and expression of genes encoding colonization factors that favor the selection and persistence of these strains in hospitals.
Introduction
Klebsiella pneumoniae belongs to the Enterobacteriaceae family. It is a non-motile, Gram-negative, and facultative anaerobic bacilli. K. pneumoniae resides on plants, soil, and water and exists as part of the normal microbiota of the nasopharynx and gastrointestinal tract of humans. K. pneumoniae is considered one of the most important opportunistic pathogens associated with nosocomial and community-acquired infections, particularly in hospitalized immunocompromised individuals; likewise, infections produced by K. pneumoniae increase with the indiscriminate use of antimicrobial agents.1K. pneumoniae mainly causes infections in burn wounds and respiratory and urinary tracts, although it has also been recently associated with liver abscesses in Asian countries due to the K1 and K2 capsular serotypes.1,2 The pathogenicity of K. pneumoniae is mediated by several virulence factors that allow it to evade host innate immune responses. These factors include the capsule, lipopolysaccharide, adhesins, iron acquisition systems, resistance to serum, and biofilm formation.1,3,4 Types 1 and 3 fimbriae are produced and assembled on the surface of clinical isolates of K. pneumoniae.5 Type 1 fimbria is the best characterized, highly prevalent in many species of the Enterobacteriaceae family, and is mainly composed of several structural subunits called FimA. FimH, an adhesin that confers the ability to recognize mannose, is located at the tip of the fimbria interacting with FimA.6 Type 3 fimbriae are characterized by the agglutination of erythrocytes treated with tannic acid, mediate bacterial adherence to endothelial and bladder cell lines and are involved in biofilm formation on abiotic surfaces.4,7 Some strains of K. pneumoniae possess plasmids that code for extended spectrum-type beta-lactamase (ESBL) enzymes, which promote resistance to β-lactam antibiotics and are associated with the failure of treatments and high morbidity and mortality rates.8,9 Although K. pneumoniae is one of the major opportunistic pathogens, additional studies are needed to characterize the strains involved in hospital outbreaks and the associated virulence factors. The aim of this study was to characterize 50 clinical isolates of K. pneumoniae isolated from urine, blood, cerebrospinal fluid, and catheters based on their resistance to antibiotics and the presence of virulence factors associated with their persistence in the hospital environment.
Subjects and methods
Inclusion criteria
All patients from 0-18 years of age who were treated at the Hospital Infantil de México Federico Gómez (HIMFG) between January 1 and December 31 2009 were considered for analysis. Relevant K. pneumoniae clinical isolates obtained from blood, urine, cerebrospinal fluid, and catheters were included in our analysis.
Biochemical identification
Phenotypic identification of each isolate was performed based on conventional biochemical tests: TSI (triple sugar-iron, Bioxon, México), LIA (lysine-iron agar, Bioxon), MIO (motility medium-indole-ornithine, Bioxon®, urea in Christensen base (Bioxon), Simmons citrate (Bioxon), methyl red, and Voges-Proskauer (Bioxon).
Antibiotic susceptibility
Antibiotic susceptibility testing was performed by the Kirby-Bauer method according to the Clinical Laboratory Standards Institute (CSLI).10 A colony from each K. pneumoniae strain was grown overnight in Mueller Hinton broth (Becton Dickinson, USA) at 37 ºC. Bacterial cultures were adjusted to 0.5 on the MacFarland nephelometer scale (1.5 x 108 CFU/ml) and plated on Mueller Hinton agar (MCD LAB, Mexico) by the streaking method using a sterile swab. The antibiotics (Becton Dickinson) evaluated included cefpodoxime (CPD) 10 μg, ceftazidime (CAZ) 30 μg, cefepime (FEP) 30 μg, ceftriaxone (CRO) 30 μg, aztreonam (ATM) 30 μg, gentamicin (GM) 10 μg, amikacin (AK) 30 μg, ciprofloxacin (CIP) 5 μg, ofloxacin (OFX) 10 μg, meropenem (MEM) 10 μg, imipenem (IMP) 10 μg, and trimethoprim/sulfamethoxazole (SXT) 1.25/23.75 μg. E. coli ATCC 25922 and K. pneumoniae ATCC 700603 strains were used as controls.
Presumptive phenotypic test for the detection of extended spectrum β-lactamases
The test for the detection of extended spectrum β-lactamases was based on the use of ceftazidime/clavulanic acid (30/10 μg) and their interaction with aztreonam (30 μg) and ceftazidime (30 μg) according to the criteria from the CLSI.10E. coli ATCC 25922 and K. pneumoniae ATCC 700603 strains were used as controls.
Phenotypic confirmatory test for extended spectrum b-lactamases detection
The ESBL confirmation test was performed using ceftazidime (30 μg) and ceftazidime/clavulanic acid (30/10 μg) according to the criteria suggested by the CLSI.10 The ESBL confirmation test was considered positive when the zone of inhibition observed presented a difference of ≥5 mm on the ceftazidime disk relative to the ceftazidime disk with clavulanic acid. E. coli ATCC 25922 and K. pneumoniae ATCC 700603 strains were used as controls.
Biofilm assays
Quantification of biofilm formation by K. pneumoniae was performed according to the method published by O'Toole and Kolter in 1998.11 Briefly, a colony from each of the K. pneumoniae strains was grown in brain-heart infusion broth (BHI, Becton Dickinson) at 37 ºC for 18 h. From this culture, 20 μl of a bacterial suspension of 0.5 MacFarland standard (1.5 x 108 CFU/ml) was used to inoculate 96-well polystyrene plates (Costar, USA) containing 180 μl of BHI, and the plates were incubated at 37 ºC for 24 h. Subsequently, the medium was removed from the plates and the wells were washed three times with 1X PBS and fixed with 2% formalin at 4 ºC for 1 h. Immediately, the samples were stained with 200 μl of 1% crystal violet for 20 min (Fluka Analytical, USA). Excess dye was removed, and 200 μl of methanol (Fermont, Mexico) was added to the wells. Color intensity was determined using a Multiskan FC (Thermo Scientific, Waltham, MA) at a wavelength of 620 nm. K. pneumoniae ATCC 700603 strain was used as positive control, and an E. coli K-12 was used as a negative control.
Visualization of K. pneumoniae fimbriae
Clinical strains of K. pneumoniae were inoculated into Luria Bertani broth (LB, Becton Dickinson) and incubated at 37 ºC for 24 h. Subsequently, the bacterial culture was centrifuged at 3000 rpm for 5 min. The cell pellet was resuspended in 1X phosphate-buffered saline (PBS), and the bacterial samples were mounted on copper grids (200 mesh and 3.05 mm diameter). The samples were stained with phosphotungstic acid solution and visualized by transmission electron microscope (TEM) at 60 kV (JEOL JEM-1010 Electron Microscope, Peabody, MA).
Adherence assays
HT-29 and A549 cell lines were cultured in T75 cm2 flasks in high glucose Dulbecco's Modified Eagle's medium (DMEM, Invitrogen, Carlsbad, CA) supplemented with 10% inactivated fetal bovine serum (FBS, Gibco, Grand Island, NY) incubated at 37 ºC in an atmosphere of 5% CO2. The cells were detached with 0.25% trypsin (Gibco) and resuspended in 24 ml of DMEM. Subsequently, 1 ml of the cell suspension was placed in each well of a 24-well culture plate (12 mm diameter), and incubated at 37 ºC in an atmosphere of 5% CO2 for 24 h. Cell monolayers with 80% confluence (2.5 × 106 cells/ml) were infected with 50 ml of the bacterial suspension (1 x 108 bacteria/ ml) and incubated at 37 ºC for 3 h in an atmosphere of 5% CO2. After 3 h of infection, the cells were washed three times with 1X PBS (pH 7.4) and incubated with 1 ml of 0.1% Triton-100X (Amresco Bioscience, Sweden) for 5 min. Subsequently, serial dilutions were plated on trypticase soy agar and incubated at 37 ºC for 24 h. Adhesion assays were performed in triplicate to obtain an average of the colony forming units (CFU/ml).
Detection of the ecpA, fimH, and mrkA genes
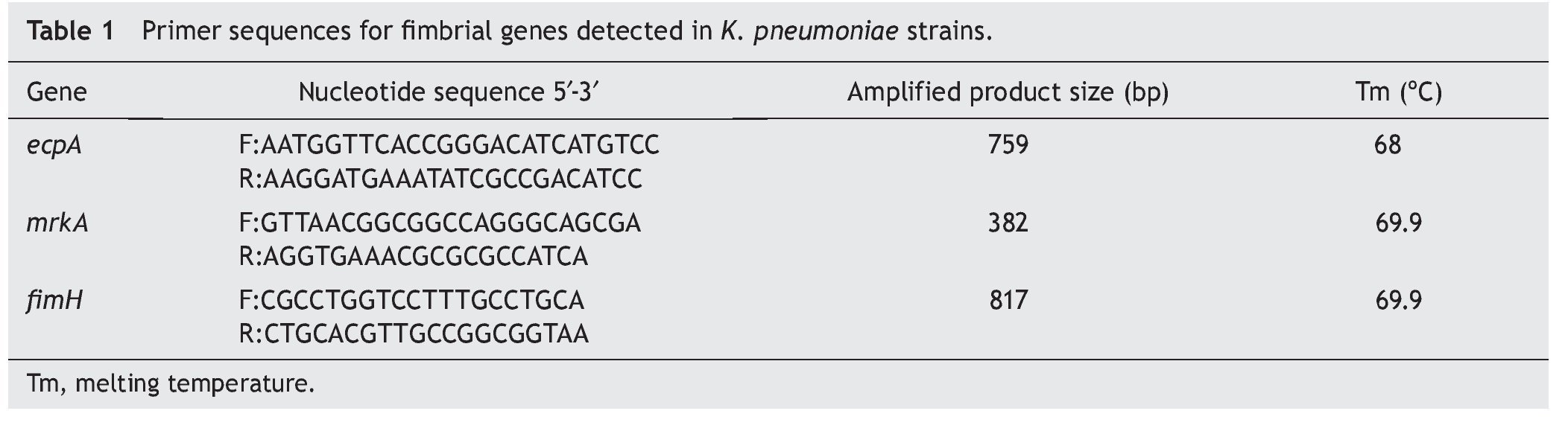
The ecpA, fimH, and mrkA genes were detected based on the published genome of K. pneumoniae strain NTUH-K2044. The nucleotide sequences obtained for each gene were analyzed using the Basic Local Alignment Search Tool (BLAST) program. The sequence with an appropriate identity "score" was used to design primers with the Primer-BLAST program. PCR assays were performed using a commercial PCR kit (Promega, Madison, WI) with the following reaction mixtures: 5 μl of reaction buffer [MgCl2 (50 mM), Taq polymerase, and dNTPs (2 mM)], 0.5 μl of primer F (10 μM), 0.5 μl of primer R (10 μM), 3 μl of water, and 1 μl of DNA (100 ng) for each strain. PCR reactions were conducted in a thermal cycler (Applied Biosystems GeneAmp®PCR System 9700, Foster City, CA) (Table 1). PCR products were run in a 1.5% agarose gel with TAE (Tris-Acetate-EDTA) at 100 V in an electrophoretic chamber (APOLLO Instrumentation, Continental Lab Products, Mexico). Gels were stained with ethidium bromide (10 μg/ml) and visualized by UV light (Bio-Imaging Systems, AccesoLab, Mexico). The genomic DNA of strain K. pneumoniae ATCC 700603 was used as a positive control, whereas one from E. coli HB101 and E. coli EDL933 were used as negative controls.
Expression of ecpA, fimH, and mrkA genes in K. pneumonia
Quantification of the expression of the three genes (ecpA, fimH, and mrkA) was performed by RT-PCR. The genomic cDNA of the K. pneumoniae ATCC 700603 strain was used as a positive control, and the ribosomal 16S gene was used as an internal control.12 All strains used were grown in LB broth and incubated at 37 ºC for 18 h. RNA was purified by using the RNeasy commercial kit (Qiagen, USA) and was treated with DNase to remove any contaminants (Invitrogen). Using 1 μg/ml of purified RNA as a template, cDNA was obtained using a Reverse Transcription System kit (Promega) and a reaction mixture [(MgCl2 (25 mM) buffer, dNTPs (10 mM), AMV reverse transcriptase, random oligo, RNA (1 μg/μl), and nuclease-free water]. The cDNA obtained was diluted in nuclease-free water and stored at -70 ºC.
The PCR reaction was carried out using the commercial Reverse Transcription-PCR kit (Promega) with the following reaction mixture: MgCl2 (25 mM), recombinase enzyme (10 mM), dNTPs (10 mM), primers (10 μM, F and R primer), 5 μl of nuclease-free water, and 5 μl of cDNA (200 ng) from K. pneumoniae ATCC 700603 strain or from the clinical strains. The PCR reaction was performed in a thermocycler (Applied Biosystems GeneAmp® PCR System 9700, Foster City, CA) (Table 1). The products obtained by RT-PCR were viewed in 1.5% agarose gels, ran in TAE buffer at 100 V, stained with ethidium bromide (10 μg/ml), and then visualized by UV light (Bio-Imaging Systems). The expression of the genes was determined according to the intensity of the amplified product in each of the samples.
Results
Fifty K. pneumoniae strains isolated from 34 patients hospitalized in HIMFG with urinary tract infections, asymptomatic bacteriuria, bacteremia, sepsis, or ventriculitis associated with ventilation systems from January to December 2009 were included in our analysis. Forty-two percent (14/34) of the patients acquired nosocomial infections, 42% (14/34) presented sepsis, and 18% (6/34) developed septic shock. In addition, K. pneumoniae strains were isolated from 48% (24/50) of urine cultures, 24% (12/50) of blood, 18% (9/50) of catheters, and 10% (5/50) of cerebrospinal fluid sources.
Antibiotic resistance observed in the K. pneumoniae isolates
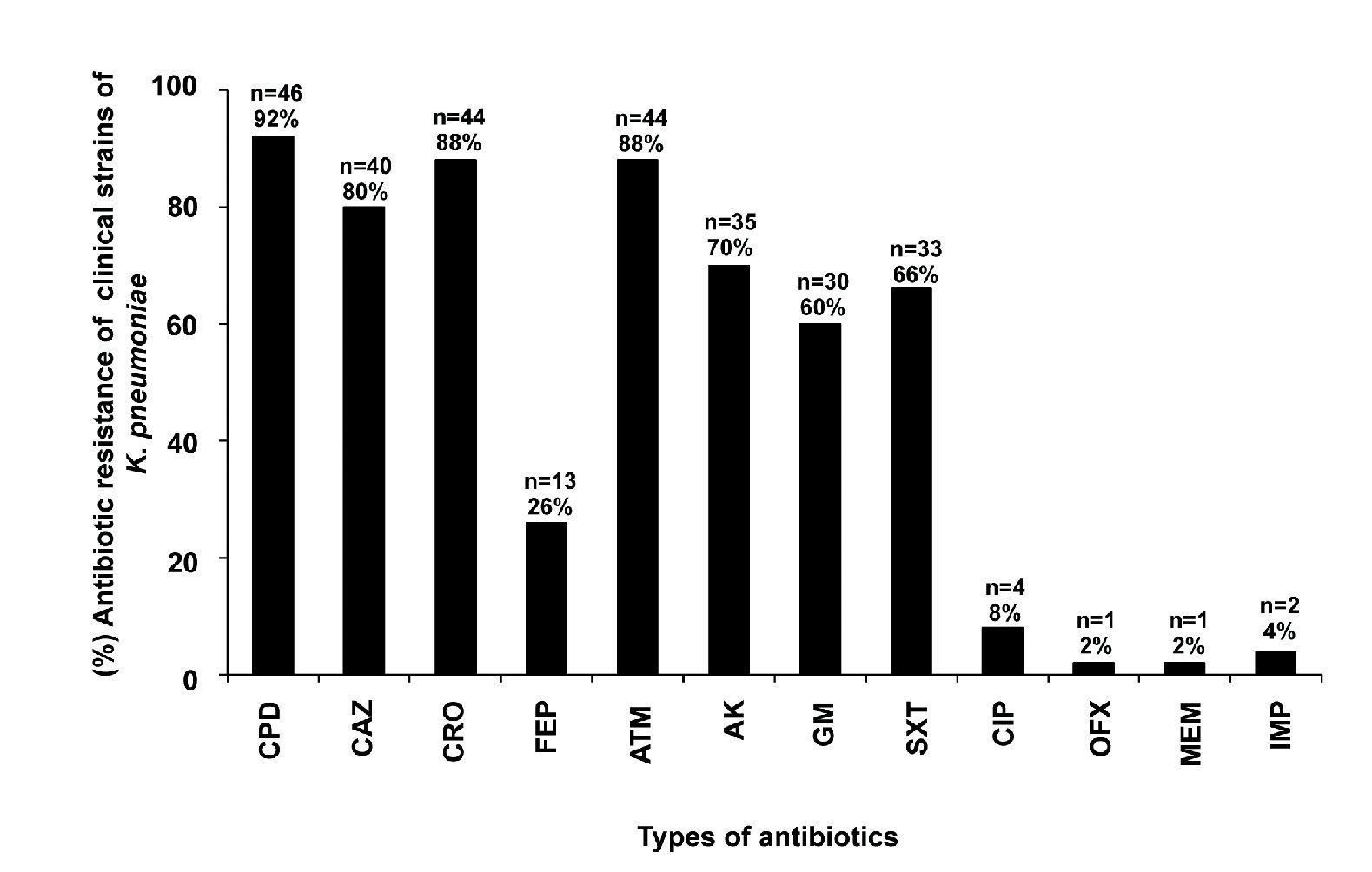
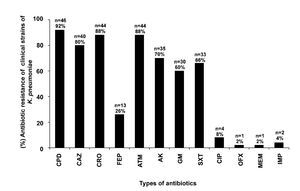
Some of the clinical strains of K. pneumoniae showed resistance to CPD (92%, 46/50), CRO (88%, 44/50), CAZ (80%, 40/50), FEP (26%, 13/50), ATM (88%, 44/50), AK (70%, 35/50), GM (60%, 30/50), SXT (66%, 33/50), CIP (8%, 4/50), OFX (2%, 1/50), MEM (2%, 1/50), and IMP (4%, 2/50) (Fig. 1). Eighty-eight percent (44/50) of the K. pneumoniae clinical isolates were resistant to at least three families of antibiotics (data not shown).
Figure 1 Resistance test to 12 antibiotic families of K. pneumoniae strains isolated from different clinical sources. Cefpodoxime (CPD), ceftazidime (CAZ), ceftriaxone (CRO), cefepime (FEP), aztreonam (ATM), amikacin (AK), gentamicin (GM), trimethoprim/ sulfamethoxazole (SXT), ciprofloxacin (CIP), ofloxacin (OFX), meropenem (MEM), and imipenem (IMP).
The presumptive test for ESBL production was positive for 90% (41/46) of the strains resistant to third-generation cephalosporins. Of the strains that were positive for the presumptive ESBL test, the ESBL phenotype was further confirmed in 97% (40/41) of the strains (data not shown).
Biofilm formation in K. pneumoniae clinical strains
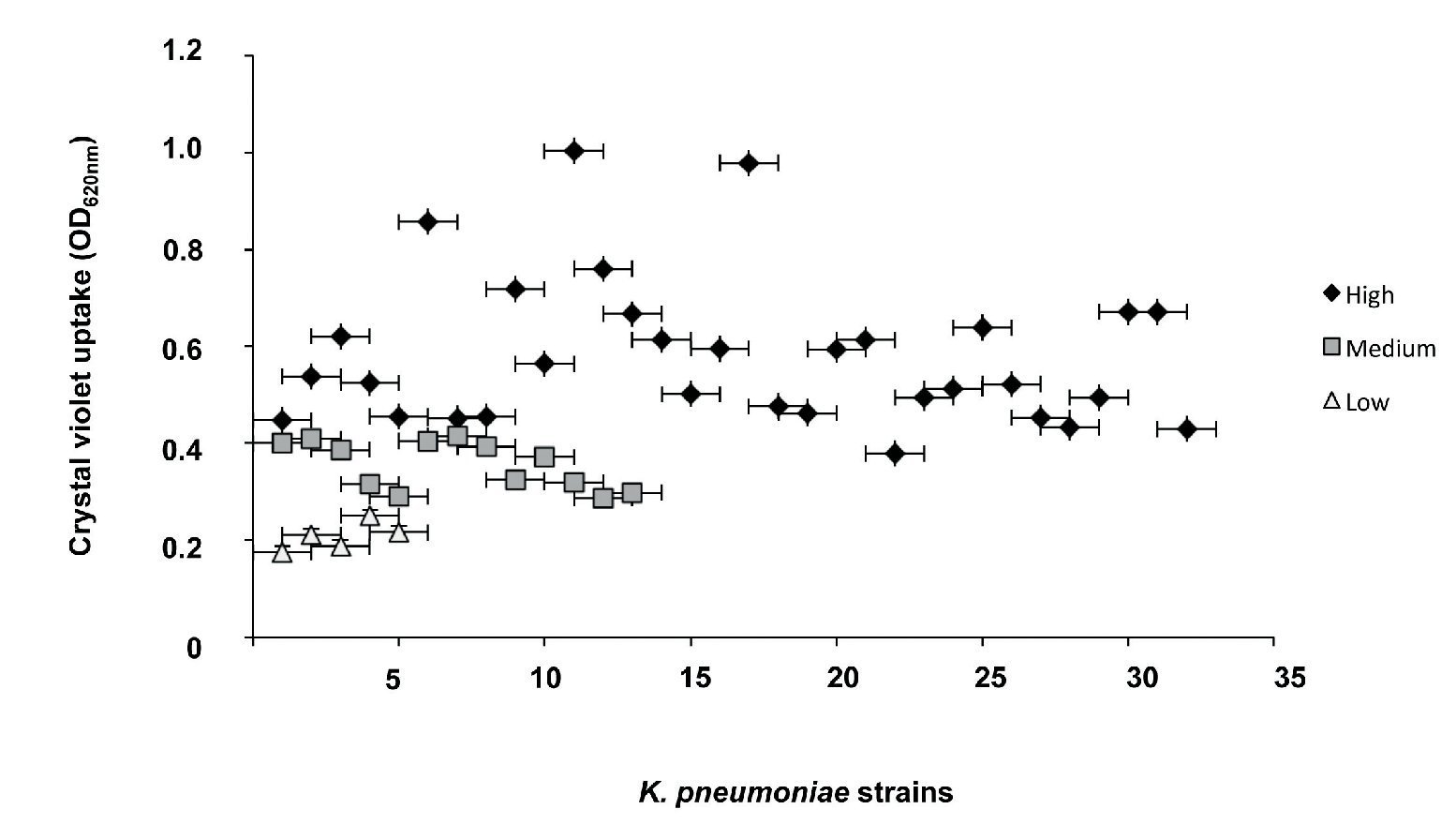
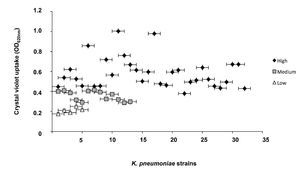
The classification of biofilm-producing strains was established based on the absorbance readings obtained from the ratio of the blank (0.069) and the positive control (0.744). Strains with absorbance readings <0.138 were considered non-biofilm formers. Absorbance values between two and four times the reading of the blank (0.139-0.276) were considered low biofilm-forming strains. Values between four and six times the blank reading (0.277-0.414) were considered medium biofilm-forming strains, whereas strains with an absorbance greater than six times that of the blank (>0.415) were classified as high biofilm formers. Our analysis showed that 100% of the K. pneumoniae strains were able to form a biofilm. Furthermore, 64% (32/50) of the strains were high biofilm formers, 26% (13/50) were medium biofilm formers, and 10% (5/50) were low biofilm formers (Fig. 2).
Figure 2 Quantitative analysis of biofilm formation on abiotic surface of clinical strains of K. pneumoniae. Absorbance values of 0.139 to 0276 at OD600nm were considered as low biofilm producers, values of 0.277 to 0.414 medium biofilm producer, and values >0.415 high biofilm producers.
Visualization of capsule and fimbriae in K. pneumoniae strains
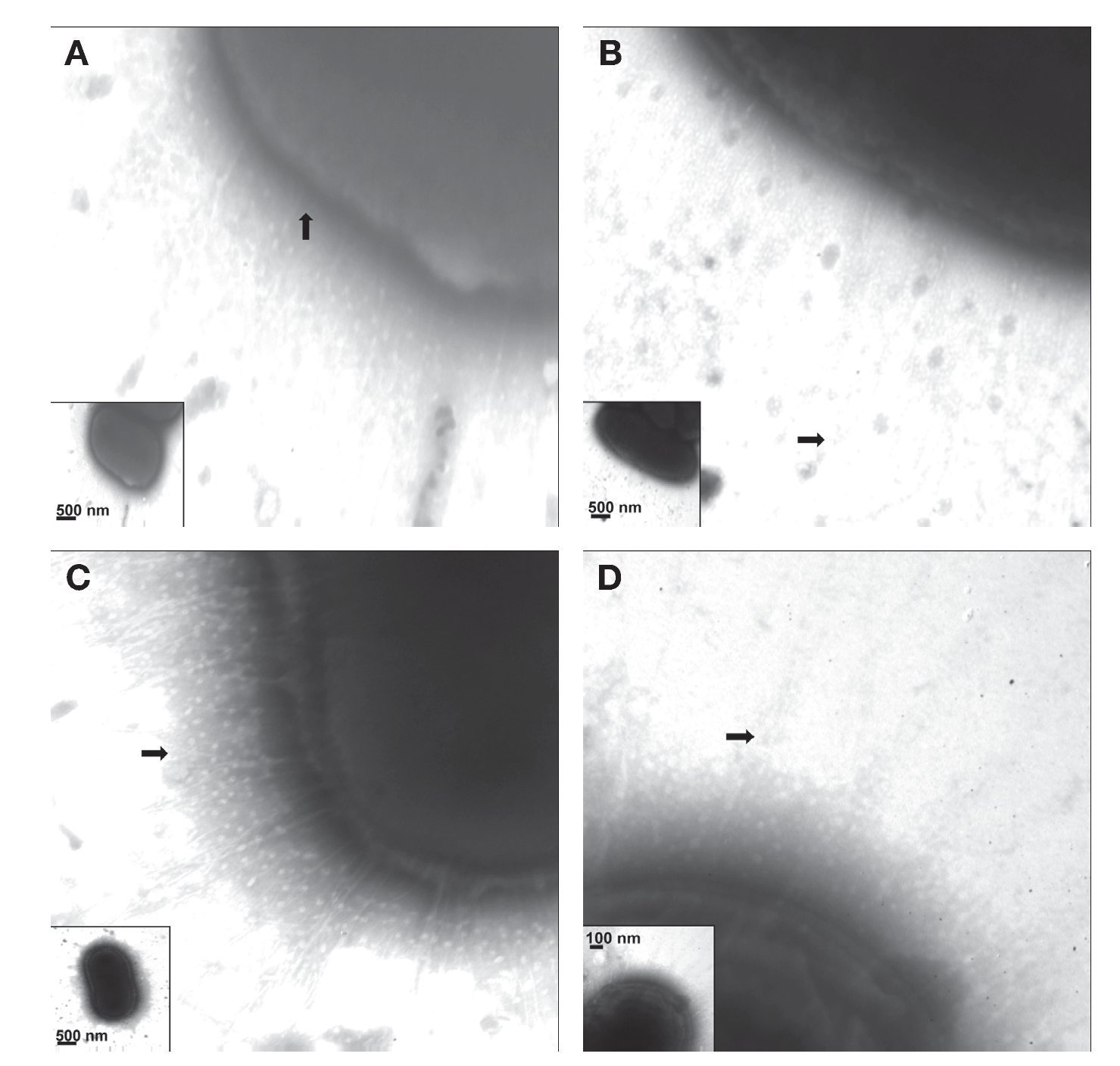
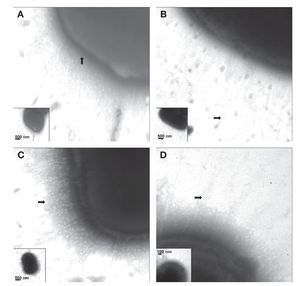
Fimbriae of K. pneumoniae strains stained with phosphotungstic acid were visualized by TEM. The electron micrographs showed the presence of capsular structures as a prominent feature in the morphology of K. pneumoniae (Fig. 3A). Urine source produced more abundant and longer fimbriae with lengths of ∼700-900 nm (Fig. 3B). Meanwhile, blood strains showed less abundant fimbrial structures with an average length of 200-400 nm (Fig. 3C). Fimbriae visualized in cerebrospinal fluid strains showed an approximate length of 350-500 nm (Fig. 3D) and the fimbria from K. pneumoniae isolated from catheters exhibited an approximate length of 300-650 nm.
Figure 3 Visualization of capsule and fimbriae by TEM. (A) Clinical strains of K. pneumoniae isolated from cerebrospinal fluid source showing capsular structure when grown in LB and at 37 ºC. (B-D) Clinical strains of K. pneumoniae isolated from urine, cerebrospinal fluid, and bloodstream producing different types of fimbriae at the cell surface. Samples were negatively stained and electron micrographs were taken at a magnification of 40,000× (A and C), 50,000× (B), and 75,000× (D). The whole bacteria are indicated in the inserts.
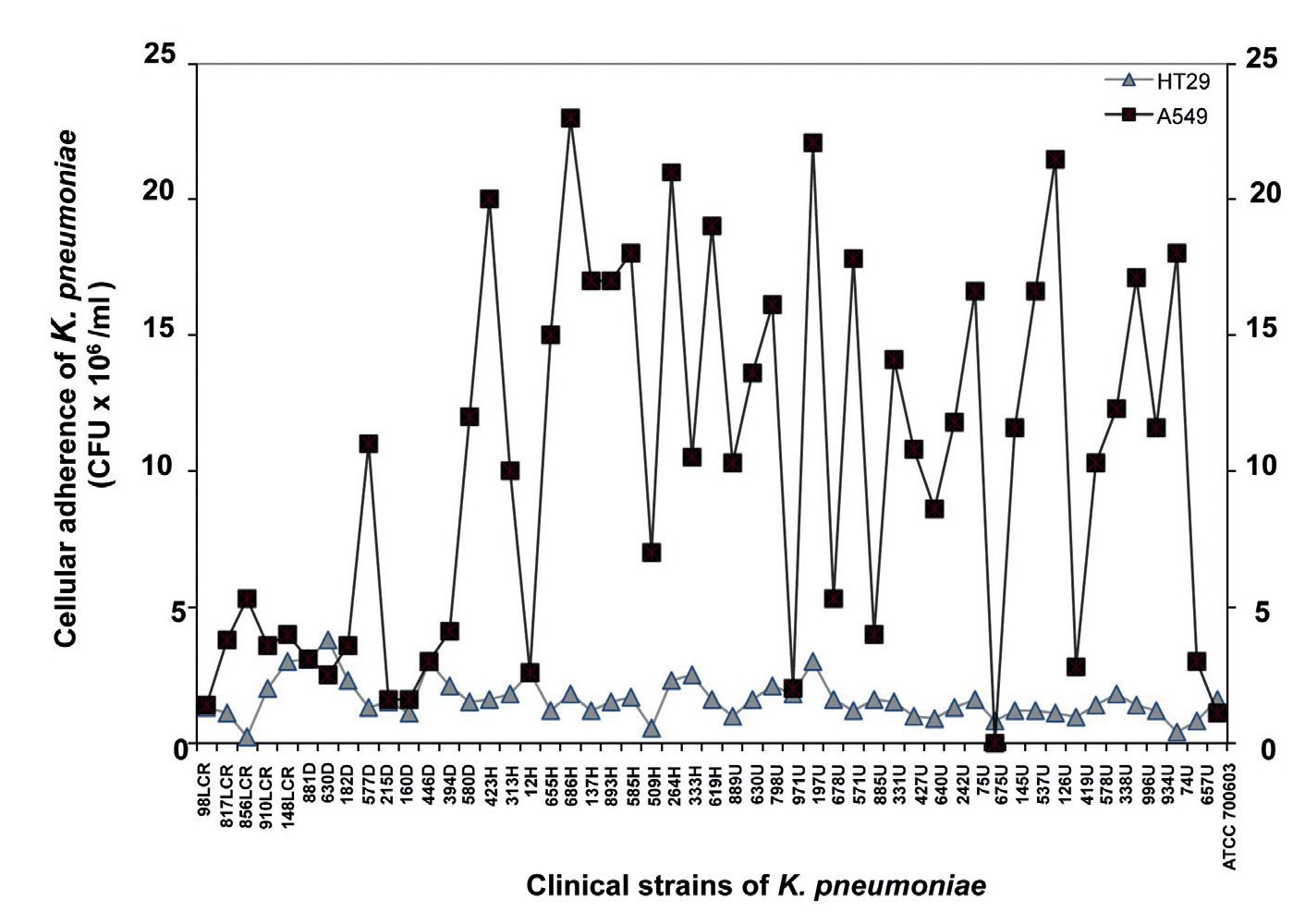
Adherence to A549 and HT29 cell lines
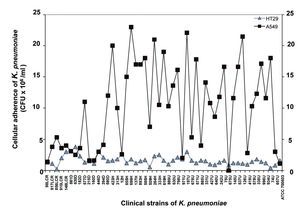
Quantitative analysis of adhesion assays using A549 cells showed a minimum value of 0.11 x 106 CFU/ml and a maximum value of 23 x 106 CFU/ml, whereas adhesion assays using HT-29 cells produced a minimum value of 0.23 x 106 CFU/ml and a maximum value of 3.8 x 106 CFU/ml (Fig. 4). Based on the quantitative analysis observed in both cell lines, strains of K. pneumoniae showed higher adherence to the A549 cell line relative to the HT-29 cell line (Fig. 4). These data suggest that A549 adenocarcinoma human alveolar basal epithelial cells are more susceptible to infection by K. pneumoniae than intestinal HT-29 cells.
Figure 4 Quantification of bacterial adherence to human epithelial cells. The clinical strains of K. pneumoniae isolated from different clinical sources were incubated with human A549 and HT-29 cells for 4 h and the adherent bacteria expressed as CFU after plating serial dilutions. Data are representative of at least three experiments performed in triplicate.
Genes associated with colonization factors in K. pneumonia
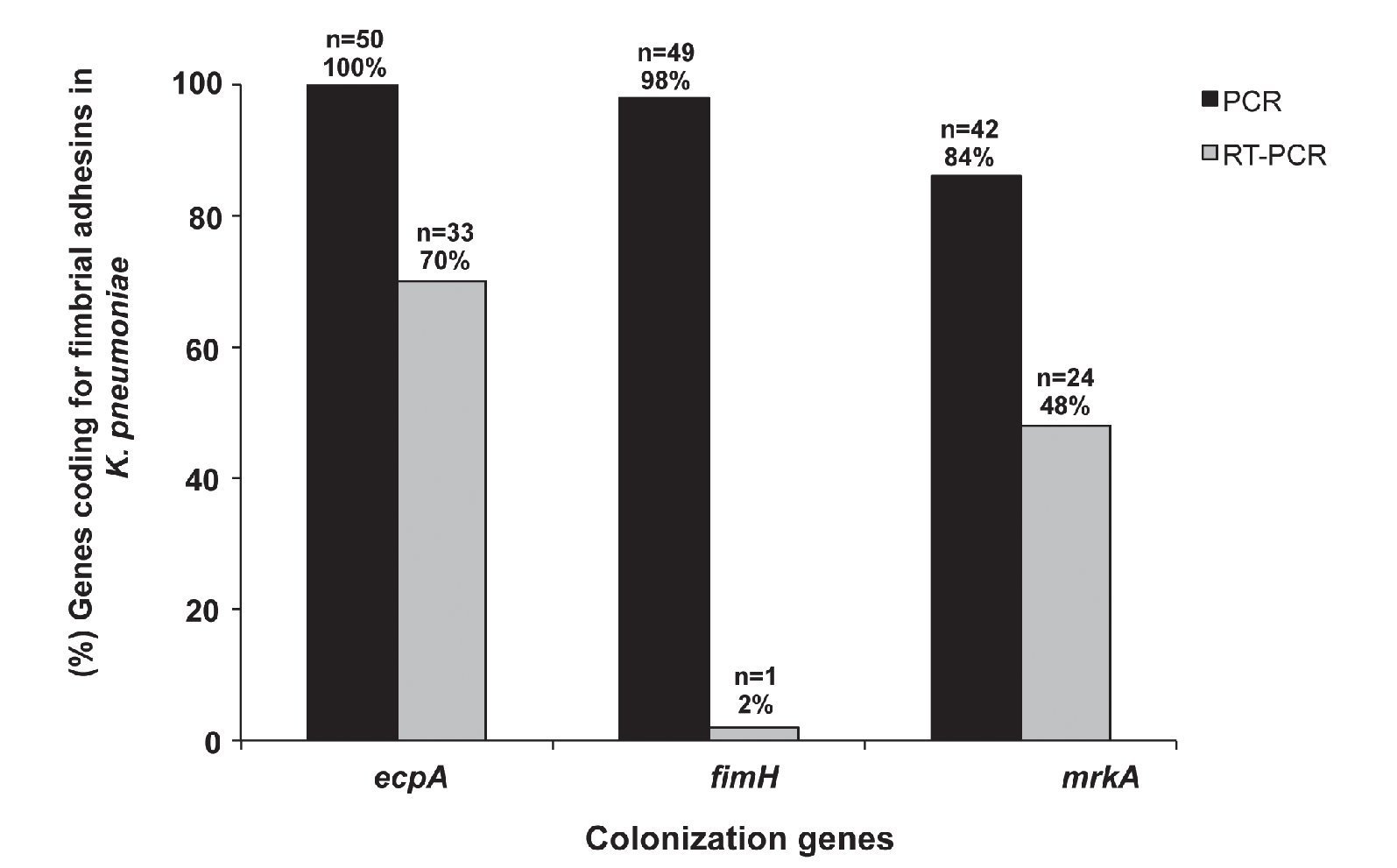
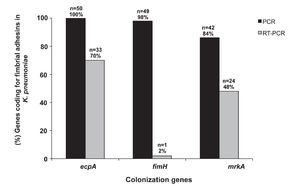
Our results show that 100% (50/50) of the clinical strains of K. pneumoniae amplified a 759-bp product corresponding to the molecular weight of the ecpA gene. Moreover, 84% (42/50) of the strains amplified the mrkA gene corresponding to a 382-bp product, and 98% (49/50) amplified the fimH gene corresponding to an 817-bp product (Fig. 5). Interestingly, 84% (42/50) of the strains possessed ecpA+/fimH+/mrkA+ genotype, 14% (7/50) ecpA+/fimH+/ mrkA- genotype, and 2% (1/50) of the strains the ecpA+/ fimH-/mrkA+ genotype.
Figure 5 Detection (PCR) and expression (RT-PCR) of ecpA, fimH, and mrkA of clinical strains of K. pneumoniae isolated from different sources.
Expression of ecpA, fimH, and mrkA genes was determined by endpoint RT-PCR. The results showed that 70% (33/50) of the strains amplified a 759-bp product consistent with the expression of the ecpA gene, 48% (24/50) amplified a 382-bp product consistent with the mrkA gene, and 2% (1/50) amplified an 817-bp product consistent with the fimH gene (Fig. 5). The 16S rRNA gene was used as an internal control showing a 162-bp product in all of the clinical strains.
Discussion
K. pneumoniae is an important pathogen in the hospital environment often associated with nosocomial infections in intensive care units and pediatric wards. In recent years, the frequency of K. pneumoniae strains resistant to third-generation cephalosporins with combined resistance to multiple antibiotics has increased. The spread of ESBL and carbapenemases among the Enterobacteriaceae family including K. pneumoniae is considered a worldwide public health problem.
In this study, 50 clinical strains of K. pneumoniae isolated from 34 patients between 0 and 18 years of age were characterized from urine (48%), blood (24%), catheters (18%), and cerebrospinal fluid sources (10%). Of the different types of clinical sources analyzed, urine most commonly yielded Klebsiella strains, which is consistent with a previous report by Stahlhut et al.13 Our results showed that K. pneumoniae strains have a high degree of resistance to third-generation cephalosporins (92%) and a relatively low degree of resistance to carbapenems and quinolones. Generally, cephalosporins are used as first-line therapy for urinary tract infections and septicemia. Therefore, the indiscriminate use of cephalosporins for the treatment of such infections has resulted in the selection of resistant strains in hospitals worldwide.14
In this study, the antibiotic resistance tests indicated that 88% (44/50) of K. pneumoniae isolates were MDR, suggesting that the treatment of K. pneumoniae infections can be difficult. The high percentage of MDR strains showed an association with ESBL-positive strains (97%), although previous reports have suggested that ESBL-positive strains are generally resistant to other antibiotics.15 Infections caused by ESBL-positive strains with resistance to other antibiotics are associated with high morbidity and mortality rates. Therefore, the epidemiological surveillance of these strains remains important.16-19
Biofilm-forming clinical strains are associated with the ability to survive in hospital environments, on medical implants, and in patient surgical wounds.20-22 We observed that 100% (50/50) of K. pneumoniae strains formed biofilms to varying degrees. These data correlate with other recent studies that showed that 76% (41/54) of K. pneumoniae strains formed biofilms.23
In Gram-negative bacteria such as Acinetobacter baumannii, previous studies have shown that biofilm-forming strains are resistant to aminoglycosides, carbepenems, tetracyclines, and sulfonamides.24 In K. pneumoniae and Staphylococcus aureus, the ability to form biofilms has been observed in strains resistant to cephalosporins and fluoroquinolones, respectively.25 The ability to form a biofilm was associated with K. pneumoniae clinical strains that were resistant to cephalosporins, aminoglycosides, and folate inhibitors/sulfonamides. However, the ability of MDR strains to form extensive biofilms, which promotes antimicrobial resistance through the selection of highly resistant strains, has not been shown.24,26,27
In this study, 61% (27/44) of K. pneumoniae isolates were MDR and able to form large biofilms, which are important pathogenic features because K. pneumoniae can cause sepsis in immunocompromised patients, although this type of infection is often due to catheters contaminated with the bacterium. Biofilm is an important mechanism of K. pneumoniae to adhered to catheter surfaces by different structures such as exopolisacarides, outer membrane proteins, and fimbriae.28 Moreover, factors such as bacterial adhesion and biofilm formation play an important role in the establishment of infections by K. pneumoniae.29,30
The main virulence factors associated with the pathogenesis of K. pneumoniae are type 1 fimbriae, type 3 fimbriae, and capsule.30,31 The capsule of K. pneumoniae is a structure that provides protection against bacterial phagocytosis and the bactericidal activity of serum and thus promotes nosocomial infections in the respiratory and urinary tracts. Based on the results obtained from our TEM analysis, clinical strains of K. pneumoniae isolated from HIMFG patients showed a polymeric capsule similar to previous reports for this pathogen.32 TEM results showed that fimbrial structures of different lengths, which are produced according to the source of the clinical isolate of K. pneumoniae, play an important role in the pathogenesis of K. pneumoniae.
Characterization of the K. pneumoniae strains was based on their ability to adhere to cells and express genes that coding for three types of fimbriae. The quantitative analysis of the adhesion assays of the clinical strains of K. pneumoniae showed a greater affinity for the A549 cell line with respect to the HT-29 cell line. These results suggest a high correlation between the level of K. pneumoniae adhesion and the A549 cells derived from a lung carcinoma with the ability to express specific receptors that mediate the interaction with K. pneumoniae. Tarkkanen et al. showed that type 3 fimbriae bind mainly to the basolateral surface of cells of tracheal source and to basal epithelial cells of bronchial source.33
The frequency of the ecpA, fimH, and mrkA genes in the 50 clinical strains of K. pneumoniae was determined by PCR. According to the results, the presence of these genes was detected at different percentages with a frequency of 100% for ecpA, 98% for fimH, and 84% for mrkA. These results are consistent with the ubiquitous nature of these fimbriae in K. pneumoniae, as has been reported in other studies.1,34
ECP (E. coli Common Pilus) is a highly conserved fimbriae in enteric species such as Klebsiella spp. and K. pneumoniae (98% and 97%, respectively).35 ECP is peritrichously distributed in the outer membrane of the bacterium. ECP are short and semi-flexible fimbriae made up mainly by the 18-kDa EcpA adhesin. Our results showed 100% frequency of the ecpA gene in the clinical isolates analyzed, which is consistent with recently described data.34,35
Type 1 fimbriae are produced by several species of the Enterobacteriaceae32 family, including nonpathogenic strains.36 Type 1 fimbriae is an adhesive heteropolymeric structure based on the presence of the FimH adhesin, which mediates adhesion to manno-oligosaccharide receptors present on the surface of various cells.32 Interestingly, our results are consistent with data reported by Stahlhut et al., which showed >90% frequency for the fimH gene in clinical K. pneumoniae isolates from urine, blood, liver abscesses, and sputum.13 Likewise, similar percentages were detected in environmental K. pneumoniae strains isolated from plants and water. Type 1 fimbriae plays an important role in urinary tract infections caused by K. pneumoniae and is also involved in biofilm formation.37 Type 3 fimbriae confers K. pneumoniae the ability to adhere to the extracellular matrix of cells. Through the MrkA protein, type 3 fimbria mediates bacterial attachment to the basolateral surface of various cell types as the tracheal epithelium and renal tubular cells.7,38 These fimbriae are involved in biofilm formation on biotic and abiotic surfaces, which facilitates nosocomial infections in catheterized immunocompromised patients.4
Based on the results obtained by PCR, expression of the three fimbrial genes (fimH, mrkA, and ecpA) was confirmed by RT-PCR. Our results show that 70% of the clinical strains expressed ecpA, which is consistent with other studies that have shown that in a population of 169 strains of enterohemorrhagic E. coli, 71.6% expressed the ecpA gene. However, fimH gene expression in the same population of E. coli strains was only 2%, suggesting that the expression of fimbriae is related to nutritional, environmental, and osmotic stresses.34 Likewise, previous studies have suggested that expression of fimH depends on the anatomical site of bacterial colonization and the chemical and biological conditions. Nevertheless, there was no significant correlation between strains that possessed the mrkA gene and strains that expressed the mrkA gene, as 86% of the strains harbored the mrkA gene but only 48% of the strains expressed mrkA based on RT-PCR studies.
Clinical symptoms reported for the 50 K. pneumonia strains were described as urinary tract infections, asymptomatic bacteriuria, bacteremia, sepsis, and ventriculitis associated with ventilation systems. Furthermore, 42% (14/34) of patients acquired nosocomial infections by K. pneumoniae, 42% (14/34) developed sepsis, and 18% (6/34) developed septic shock. The characterization of these strains revealed that MDR was associated with a high percentage of ESBL-positive strains as well as a high percentage of the strains that formed extensive biofilms. Strains isolated from cerebrospinal fluid, catheters, and blood demonstrated strong adherence to A549 cells expressing the mrkA and ecpA genes. Furthermore, an association was observed between the ability of the strains isolated from uroculture to adhere to cells and their expression of the ecpA+/mrkA+ genes or only of the ecpA gene.
Collectively, our results suggest that clinical K. pneumoniae strains isolated from sources from children treated in the HIMFG are able to colonize biotic and abiotic surfaces, disseminate, and established in their host, resulting in the development of nosocomial infections in immunocompromised patients.
Funding
This work was supported by grant number HIM/2010/011 of Federal Funds from Hospital Infantil de México Federico Gómez.
Conflict of interest
The authors declare no conflict of interest of any nature.
Acknowledgments
We thank Dr. Briceida López Martínez for providing K. pneumoniae strains. We also thank chemist Araceli De León Ham and laboratory technician Maribel Martínez Carrión for their assistance in the identification and maintenance of the K. pneumoniae strains.
Received 24 October 2013;
accepted 12 November 2013
* Corresponding author.
E-mail address:juanxico@yahoo.com (J. Xicohtencatl-Cortes).