Nine Legionella pneumophila strains isolated from cooling towers and a standard strain (L. pneumophila serogroup 1, ATCC 33152, Philadelphia 1) were analyzed and compared in terms of motility, flagella structure, ability to form biofilms, enzymatic activities (hemolysin, nucleases, protease, phospholipase A, phospholipase C, acid phosphatase, alkaline phosphatase and lipase), hemagglutination capabilities, and pathogenicity in various host cells (Acanthamoeba castellanii ATCC 30234, mouse peritoneal macrophages and human peripheral monocytes). All the isolates of bacteria appeared to be motile and polar-flagellated and possessed the type-IV fimbria. Upon the evaluation of virulence factors, isolate 4 was found to be the most pathogenic strain, while 6 out of the 9 isolates (the isolates 1, 2, 3, 4, 5, and 7) were more virulent than the ATCC 33152 strain. The different bacterial strains exhibited differences in properties such as adhesion, penetration and reproduction in the hosts, and preferred host type. To our knowledge, this is the first study to compare the virulence of environmental L. pneumophila strains isolated in Turkey, and it provides important information relevant for understanding the epidemiology of L. pneumophila.
Legionella is the only genus in the Legionellaceae family and consists of more than 50 species and 70 serogroups. Of the 50 species, 20 are disease-causing agents. L. pneumophila is the most frequent species in the genus Legionella, and it consists of 16 serogroups.1 More than 90% of cases of Legionnaires’ disease are caused by L. pneumophila serogroup 1.
To prevent infections caused by Legionella bacteria, workplaces, particularly hotels, hospitals and factories, have their aerosol-forming units such as cooling towers that are inspected regularly for Legionella contamination and utilize cleaning and disinfection methods to prevent its colonization. In Turkey and other European countries, studies with the genus Legionella focus largely on detection and elimination. However, the presence of the bacteria in water systems does not always cause outbreaks. Whether there is an outbreak depends on various factors, such as the diversity of serogroups, inhaled bacteria count, differences in immunity in the population, and the virulence of the bacteria. Therefore, although preventing Legionella colonization in water systems is the priority, determining their virulence is also very important. With the development of molecular methods, more emphasis was given to studies investigating the pathogenicity of these bacteria. Studies on the Legionella virulence generally focus on the type II and type IV secretion systems.2,3 The type II PilD-dependent Lsp (Legionella secretion pathway) includes acid and alkali phosphatase, protease, RNase, lipase, and phospholipase A and C activities. Investigations have shown that the type II Lsp system is responsible for intracellular infection.4 The type IV secretion system of Legionella is subdivided into type IVA, which includes the Lvh (Legionella vir homologues) system, and type IVB, the dot (defective organelle trafficking)/icm (intracellular multiplication) secretory pathway. The Lvh system is responsible for host cell infection, while the dot/icm genes have an important role in intracellular replication in host cells. In addition, the type I Lss (Legionella secretion system) and a putative type V secretion system were identified in L. pneumophila.5
Studies on the pathogenicity of L. pneumophila provide a better understanding of its survival in nature, its mechanisms of host infection, the prevention of phagolysosome formation, and its mechanisms of leaving the host. The determination of virulence is obviously very important for the development of faster and more efficient treatment methods against Legionella.
Additionally, a common method of protection against Legionella outbreaks is the disinfection of water systems the bacteria colonize. However, the presence of the Legionella in the water systems has not always caused outbreaks. One of the main reasons of this is the virulence of the bacteria that are inhaled. For this reason, Legionella pathogenicity must be considered along with their presence in water system. Therefore, in the current study, the type II and IV virulence factors of different environmental isolates were determined according to L. pneumophila ATCC 33152. In this respect, it was aimed to determine the pathogenicity of Legionella colonizing water systems.
Materials and methodsBacteriaIn the current study the L. pneumophila Philadelphia 1 (ATCC 33152) standard strain and nine different L. pneumophila serogroup 1 (SG1) strains which had been isolated in previous studies from various water sources in the city of Istanbul were used. All strains were grown on BCYE (Buffered Charcoal Yeast Extract) agar medium and stored in 37% glycerol solution at −86°C.6 Throughout the study, the fresh cultures prepared from this stock were used. In this study, the bacteria were generally cultured on BCYE agar for 3 days at 37°C. However, for some enzymatic activity assays, supernatants and lysates were obtained from BYE broth cultures (post-exponential phase). The bacteria were diluted to an OD (600nm) of 0.1 in buffered yeast extract (BYE) broth and after an incubation for 2–3 days at 37°C, the bacterial growth was evaluated by measuring the optical density of the culture at 600nm (OD600) of 1. The bacteria cultures in BYE media were centrifuged for 5min at 10,000rpm at 4°C and the supernatants were separated from pellets. The supernatants were sterilized by passage through filters (0.2μm pore size). At the same time, the pellets were mixed with the same volume of lysis solution (0.1% triton X-100 and 0.2mg of lysozyme per mL) and lysed by repeated passage through a 26-gauge needle. The supernatants and lysates were either tested immediately or stored at −20°C.
Acanthamoeba castellaniiA. castellanii ATCC 30234 cultures served as inocula for the cultures in 75cm3 tissue culture bottles containing ATCC Medium 712 (2% peptone, 0.1% yeast extract, 0.1M glucose, 4mM MgSO4, 0.4mM CaCl2, 0.1% HOC(COONa)(CH2COONa)2·2H2O, 0.05mM Fe(NH4)2(SO4)2·6H2O, 2.5mM NaH2PO3, 2.5mMK2HPO3, pH 6.5) and incubated at 30°C until the amoeba developed into trophozoites. The trophozoites were stained with 1% trypan blue and counted before the experiments.
Adherence and growth in biofilm under static conditionsThe host adhesion and reproduction capability of L. pneumophila SG1 in static artificial media was investigated according to the previously published procedures.7 The bacteria grown in BCYE agar media were diluted to an OD of 0.3 at 600nm in sterile tap water. To determine adhesion properties and growth in biofilm, 96- and 24-well flat-bottom sterile polystyrene microplates were used.5 For both experiments, 200 or 1000μL bacteria suspensions in tab water were added the 96- or 24-wells microplates, respectively. While the adhesion capacities of the isolates used the tap water suspension, for determine the growth in biofilm, after attached the bacteria to polystyrene wells, tab water was carefully removed and 1000μL BYE broth was added. The number of attached cells and the extent of biofilm formation were determined using crystal violet (CV) staining. Firstly, the well contents were collected and the wells washed gently three times. Secondly the wells were filled with 1% CV at 15min. After washing steps the dye which had been absorbed by biofilm was recovered using ethanol:acetone (80:20) incubation for 15min. Recovered solutions were transferred to another microplate and quantified by spectrophotometry at 595nm. The sterile tap water was used as a negative control. The cell adhesion was evaluated according to the criteria reported by Stepanovic et al.8
Motility and flagella structureThe motility of the bacteria grown in BCYE agar media was investigated by light microscopy. The flagella structures were studied using Leifson flagella staining.9
Hemagglutination activityErythrocyte suspensions (3% in phosphate-buffered saline [PBS]) with or without 2% d-mannose addition were used to determine d-mannose-resistant hemagglutination activity.10 the fresh bacteria grown on BCYE agar were suspended in sterile tap water to a concentration of 2×106 cells per mL. One hundred microliters of the bacterial suspension was mixed with 100μL of human erythrocyte suspension (3% in PBS) on sterile slides, and the mixture was shaken by rotating circularly for 10min at room temperature. The results were scored as ++++ (strong agglutination within 2min), +++ (strong agglutination within 10min), ++ (lower agglutination within 10min), + (fine agglutination seen with 4× magnification) and − (unobservable hemagglutination within 10min).
Determination of enzymatic activityThe hemolytic, protease, nuclease, phospholipase, phosphatase and lipase activities produced by the bacteria were studied. The hemolytic enzymes and proteases were studied in fresh cultures, the nucleases were studied in culture supernatants, and the rest of the enzymes were studied in both supernatants and cell lysates prepared according to the method published by Aragon et al.11
To study hemolytic activity, fresh cultures of bacteria were prepared by inoculation in BYE agar plates containing human or sheep erythrocytes (eBYEA). The plates were kept at 37°C and 5% CO2 for 3 days before the analyses.12 Following the incubation, the hemolytic index was calculated as the ratio between the diameters of hemolytic zones and the diameters of colony. The hemolytic index represented the intensity of the hemolysin production.
The protease activity was determined using the API ZYM test system (bioMerieux, Basingstoke, Hampshire, United Kingdom). This system has included 19 enzymatic reactions (Phosphatase alkaline, Esterase (C4), Esterase lipase (C8), Lipase (C14), Leucine aminopeptidase, Valine aminopeptidase, Cystine aminopeptidase, Trypsin, Chymotrypsin, hosphatase acid, Phosphoamidase, α-Galactosidase, β-Galactosidase, β-Glucuronidase, α-Glucosidase, β-Glucosidase, β-Glucosaminidase, α-Mannosidase, α-Fucosidase). The Leucine aminopeptidase, Valine aminopeptidase, Cystine aminopeptidase, Trypsin and Chymotrypsin tests were used for the detection of proteolytic activity. The fresh bacteria cultures were suspended in sterile tab water to an optical density at OD600 of 1. After the wells were filled, all the strips were incubated at 37°C for 4h. And after one drop (30μL) of the commercial reagents ZYM A and ZYM B was added, the color reactions were read, and a numerical value ranging from 0 to 5 was assigned according to the color chart provided by the manufacturer.13
The nuclease activity was estimated by the gel documentation system analysis (Gellogic 1500 Imaging System, Kodak, Rochester, NY, USA). Culture supernatants were added to Noble agar medium plates containing 0.15% RNA or 0.1% DNA and incubated overnight at room temperature, followed by 0.01% ethidium bromide staining. RNase ONE (Promega, Wisconsin, USA) and RQ1 DNase (Promega) enzymes served as positive controls.11
The phospholipase A (PLA) activity was estimated by spectrophotometry in cell lysates and supernatants by measuring dipalmitoylphosphatidylcholine (DPPC) conversion into free fatty acids using the NEFA HR2 Kit (Wako Diagnostics, Richmond, VA, USA). In this assay, the supernatants were incubated in 20mM Tris–HCl containing 3mM sodium azide, 0.5% TritonX-100 and 5mg of DPPC per mL for 15h at 37°C. Commercial phospholipase A2 from Streptomyces violaceoruber (Sigma) was used for the positive control experiment.14
The phospholipase C (PLC) was studied in the supernatants and cell lysates by measuring p-nitrophenyl phosphorylcholine (pNPPC) conversion to p-nitrophenol (pNP) at 410nm. To detect the PLC activity, the supernatant was mixed with 1mL 50mM HEPES (pH 7.5) buffer containing 5mM CaCl2, 5mM MnCl2, 3mM sodium azide, 0.5% Triton X-100 and 2.5mM pNPPC in the ratio of 1:10 and incubated overnight at 37°C. One unit of enzyme activity was defined as the amount of enzyme producing 1nM pNP in 1min. Bacillus cereus type 4 commercial phospholipase C (Sigma) was used for the positive control experiment.15
The acid and alkaline phosphatase enzymes were investigated by measuring p-nitrophenyl phosphate (N1891 and 73737, Sigma–Aldrich Chemical Co., Inc.) conversion to pNP. Ten microliters of supernatant or lysate was added to 100μL of 50mM citric acid (pH 5 or 10 to distinguish between acid and alkaline phosphatase activities) buffer containing 7.6mM p-nitrophenyl phosphate, and then after 5h of incubation at 37°C, the samples were evaluated at 410nm. One unit of enzyme was defined as the amount of enzyme that formed 1nM pNP in 1min.11Escherichia coli type III-L alkaline phosphatase (control 1) and potato type IV-S acid phosphatase (control 2) were used for positive control experiments.
Three independent assays were employed to determine the lipase activity in cell lysates and supernatants.11 One method utilized p-nitrophenyl caprilate (p-NPC) or p-nitrophenyl palmitate (p-NPP) conversion to pNP,16 another method measured the resorufin esterase activity by measuring conversion of 1,2-dilauryl-rac-glycero-3-glutaric acid resorufin ester to resorufin (measured at 572nm),17 while the last method measured the free fatty acid production activity from 1-monopalmitoyl glycerol (1-MG) and 1,2-dipalmitoyl glycerol (1,2-DG). In all the experimental groups, lipase enzymes from Rhizopus arrhizus and Pseudomonas sp. served as positive control 1 and 2, respectively.11
Investigation of L. pneumophila–host interactionThe in vitro infection capabilities of L. pneumophila isolates were studied in various hosts such as: (i) peripheral blood mononuclear cells (PBMCs) obtained from blood samples collected from volunteers,18 (ii) peritoneal macrophages (PMs) obtained from BALB/c female mice by induction with thioglycolate (TGC) medium (the procedure was approved by the Istanbul University animal experimentation local ethics commission; decision number: 2011/87),19 and (iii) A. castellanii ATCC 30234 trophozoites. In hosts (i) and (ii), PBS was used as the buffer solution while A. castellanii (Ac) buffer (4mM MgSO4, 0.4M CaCl2, 0.1% HOC(COONa) (CH2COONa)2·2H2O, 0.05mM Fe(NH4)2(SO4)2-6H2O, 2.5mM NaH2PO3, 2.5mMK2HPO3, pH 6.5) was used in host (iii). In all the experiments, fresh the L. pneumophila suspensions were prepared in appropriate buffers, and initial host:bacteria cell ratios were adjusted to 1:100.20 To evaluate host pathogenicity, the L. pneumophila cultures were studied in terms of their (i) cellular adhesion, (ii) cellular penetration and (iii) reproduction in the host. All the tests were conducted in 24-well tissue culture plates. To determine the ability of the bacteria to attach to host cells, the bacteria-host cells mixtures were washed and lysed after an incubation for 10min. To detect entry and intracellular growth, they were incubated for 1h. After 1h, all the wells were washed and filled with fresh medium, containing 100μgmL−1 gentamicin. After an incubation for 1h, the cells were washed three times and some that were labeled for detection of entry were lysed. To detect intracellular growth, the other wells were filled with an appropriate medium and incubated for 24, 48 and 72h. Following the osmotic lysis of host cells with distilled water, the bacteria were inoculated in BCYE agar medium. With the use of appropriate buffers, microscopic slides were prepared and studied using the Gimenez method and acridine orange staining.21 Cell adhesion levels and host penetration/reproduction levels were calculated according to the methods published by Coxon et al.22 and Chang et al.,23 respectively. For negative control experiments, no bacteria were used.
Statistical analysesAll the experiments were conducted as triplicates, and standard deviations were calculated for all the numerical results. The statistical significances of the differences in the virulence properties of L. pneumophila SG1 were evaluated using Student's t test. Moreover, the relationships between the virulence factors were evaluated with the Spearman or Pearson correlation test, depending on whether the results exhibited normal or non-normal distribution.24 Statistical evaluations were done using SPSS 16.0.
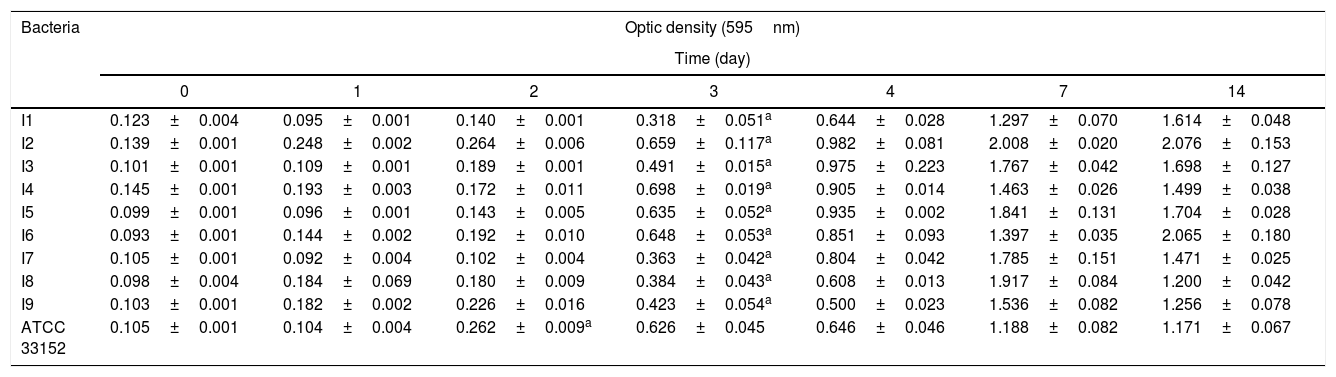
ResultsAdherence and growth in biofilm under static conditionsThe ability of L. pneumophila to adhere increased at the 6th and 24th hours of the incubation in a statistically meaningful manner (p<0.05), while there was no significant correlation between the capabilities of the isolates and of the standard L. pneumophila strain. According to Stepanovic et al.,8 the strains were classified as non-adherent (OD≤ODc), weakly adherent (ODc<OD≤2xODc), moderately adherent (2xODc<OD≤4xODc), and strongly adherent (4xODc<OD; the optical density of negative control was expressed by OD, while the cut-off OD was defined by ODc). Based upon these criteria, at the end of 6th and 24th hours, the isolate 4 appeared to adhere most efficiently, while all the isolates demonstrated statistically significant (p<0.05) increases in adhesion. Taking these results into consideration, the following assays were carried out after 6h of bacteria incubation with tap water and this point was considered as day 0. After these initial six hours of incubation, the wells were washed three times and filled with fresh BYE broth for the determination of bacterial growth in the biofilm after 1, 2, 3, 4, 7 and 14 days.
When the results were analyzed, it was determined that the biofilm growth of standard ATCC 33152 increased on the second day, while the isolates were able to reach significant (p<0.05) growth values on the third day (Table 1). At the end of the 14th day, the isolate 2 showed the highest activity according to the biofilm formation capacities of bacteria. When we statistically compared all strains to each other, there were no differences (p>0.05) between the isolates and the standard bacteria, which means that all strains had the same biofilm formation capacity.
Biofilm formation of L. pneumophila grown in static conditions in BYE broth (p:0.05).
| Bacteria | Optic density (595nm) | ||||||
|---|---|---|---|---|---|---|---|
| Time (day) | |||||||
| 0 | 1 | 2 | 3 | 4 | 7 | 14 | |
| I1 | 0.123±0.004 | 0.095±0.001 | 0.140±0.001 | 0.318±0.051a | 0.644±0.028 | 1.297±0.070 | 1.614±0.048 |
| I2 | 0.139±0.001 | 0.248±0.002 | 0.264±0.006 | 0.659±0.117a | 0.982±0.081 | 2.008±0.020 | 2.076±0.153 |
| I3 | 0.101±0.001 | 0.109±0.001 | 0.189±0.001 | 0.491±0.015a | 0.975±0.223 | 1.767±0.042 | 1.698±0.127 |
| I4 | 0.145±0.001 | 0.193±0.003 | 0.172±0.011 | 0.698±0.019a | 0.905±0.014 | 1.463±0.026 | 1.499±0.038 |
| I5 | 0.099±0.001 | 0.096±0.001 | 0.143±0.005 | 0.635±0.052a | 0.935±0.002 | 1.841±0.131 | 1.704±0.028 |
| I6 | 0.093±0.001 | 0.144±0.002 | 0.192±0.010 | 0.648±0.053a | 0.851±0.093 | 1.397±0.035 | 2.065±0.180 |
| I7 | 0.105±0.001 | 0.092±0.004 | 0.102±0.004 | 0.363±0.042a | 0.804±0.042 | 1.785±0.151 | 1.471±0.025 |
| I8 | 0.098±0.004 | 0.184±0.069 | 0.180±0.009 | 0.384±0.043a | 0.608±0.013 | 1.917±0.084 | 1.200±0.042 |
| I9 | 0.103±0.001 | 0.182±0.002 | 0.226±0.016 | 0.423±0.054a | 0.500±0.023 | 1.536±0.082 | 1.256±0.078 |
| ATCC 33152 | 0.105±0.001 | 0.104±0.004 | 0.262±0.009a | 0.626±0.045 | 0.646±0.046 | 1.188±0.082 | 1.171±0.067 |
Light microscopy examinations revealed that all the bacteria were motile and contained polar flagella.
Hemagglutination activityThe isolates 1, 2, 4 and 5 caused hemagglutination within 2min, while the isolates 6 and 7 caused hemagglutination within 10min. Isolate 2 had the fastest hemagglutination activity. All the bacteria caused mannose-resistant hemagglutination activity (MRHA) in 2min. Thus, they appeared to have type IV fimbriae.
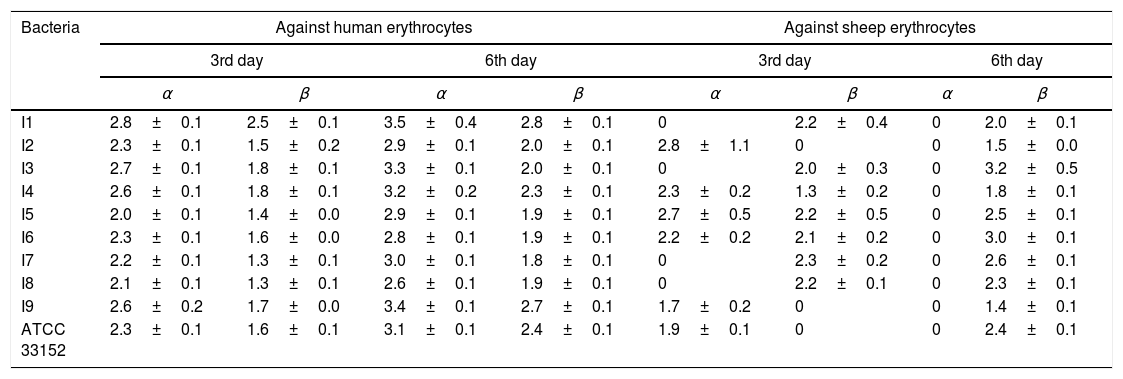
Determination of enzymatic activityStudies on human erythrocytes showed that all the examined strains had hemolytic activity (Table 2). Of all the isolates, the isolate 1 exhibited the strongest α- and β-hemolytic activity on days 3 and 6. When the hemolytic indexes were evaluated using sheep erythrocytes (Table 2), the isolate 2 had the strongest α-hemolytic activity on day 3, while isolate 7 had the strongest β-hemolytic activity on the same day. The isolate 3 exhibited the strongest β-hemolytic activity on day 6.
The hemolytic indexes of L. pneumophila strains.
| Bacteria | Against human erythrocytes | Against sheep erythrocytes | ||||||
|---|---|---|---|---|---|---|---|---|
| 3rd day | 6th day | 3rd day | 6th day | |||||
| α | β | α | β | α | β | α | β | |
| I1 | 2.8±0.1 | 2.5±0.1 | 3.5±0.4 | 2.8±0.1 | 0 | 2.2±0.4 | 0 | 2.0±0.1 |
| I2 | 2.3±0.1 | 1.5±0.2 | 2.9±0.1 | 2.0±0.1 | 2.8±1.1 | 0 | 0 | 1.5±0.0 |
| I3 | 2.7±0.1 | 1.8±0.1 | 3.3±0.1 | 2.0±0.1 | 0 | 2.0±0.3 | 0 | 3.2±0.5 |
| I4 | 2.6±0.1 | 1.8±0.1 | 3.2±0.2 | 2.3±0.1 | 2.3±0.2 | 1.3±0.2 | 0 | 1.8±0.1 |
| I5 | 2.0±0.1 | 1.4±0.0 | 2.9±0.1 | 1.9±0.1 | 2.7±0.5 | 2.2±0.5 | 0 | 2.5±0.1 |
| I6 | 2.3±0.1 | 1.6±0.0 | 2.8±0.1 | 1.9±0.1 | 2.2±0.2 | 2.1±0.2 | 0 | 3.0±0.1 |
| I7 | 2.2±0.1 | 1.3±0.1 | 3.0±0.1 | 1.8±0.1 | 0 | 2.3±0.2 | 0 | 2.6±0.1 |
| I8 | 2.1±0.1 | 1.3±0.1 | 2.6±0.1 | 1.9±0.1 | 0 | 2.2±0.1 | 0 | 2.3±0.1 |
| I9 | 2.6±0.2 | 1.7±0.0 | 3.4±0.1 | 2.7±0.1 | 1.7±0.2 | 0 | 0 | 1.4±0.1 |
| ATCC 33152 | 2.3±0.1 | 1.6±0.1 | 3.1±0.1 | 2.4±0.1 | 1.9±0.1 | 0 | 0 | 2.4±0.1 |
L. pneumophila demonstrated protease activities at leucine, valine and cystine substrates; however, they did not metabolize trypsin and chymotrypsin. The weakest protease activity was exhibited by the isolate 2 which was not able to cleave at valine and cystine substrates. The strongest enzymatic activity was found in the isolate 6.
The L. pneumophila supernatants formed much smaller nuclease zones with respect to the positive control. The isolate 1 had the highest DNase activity (20-mm zone diameter) and RNase activity (24-mm zone diameter) while the isolate 8 had the weakest DNase and RNase activities (12- and 10-mm zone diameters, respectively).
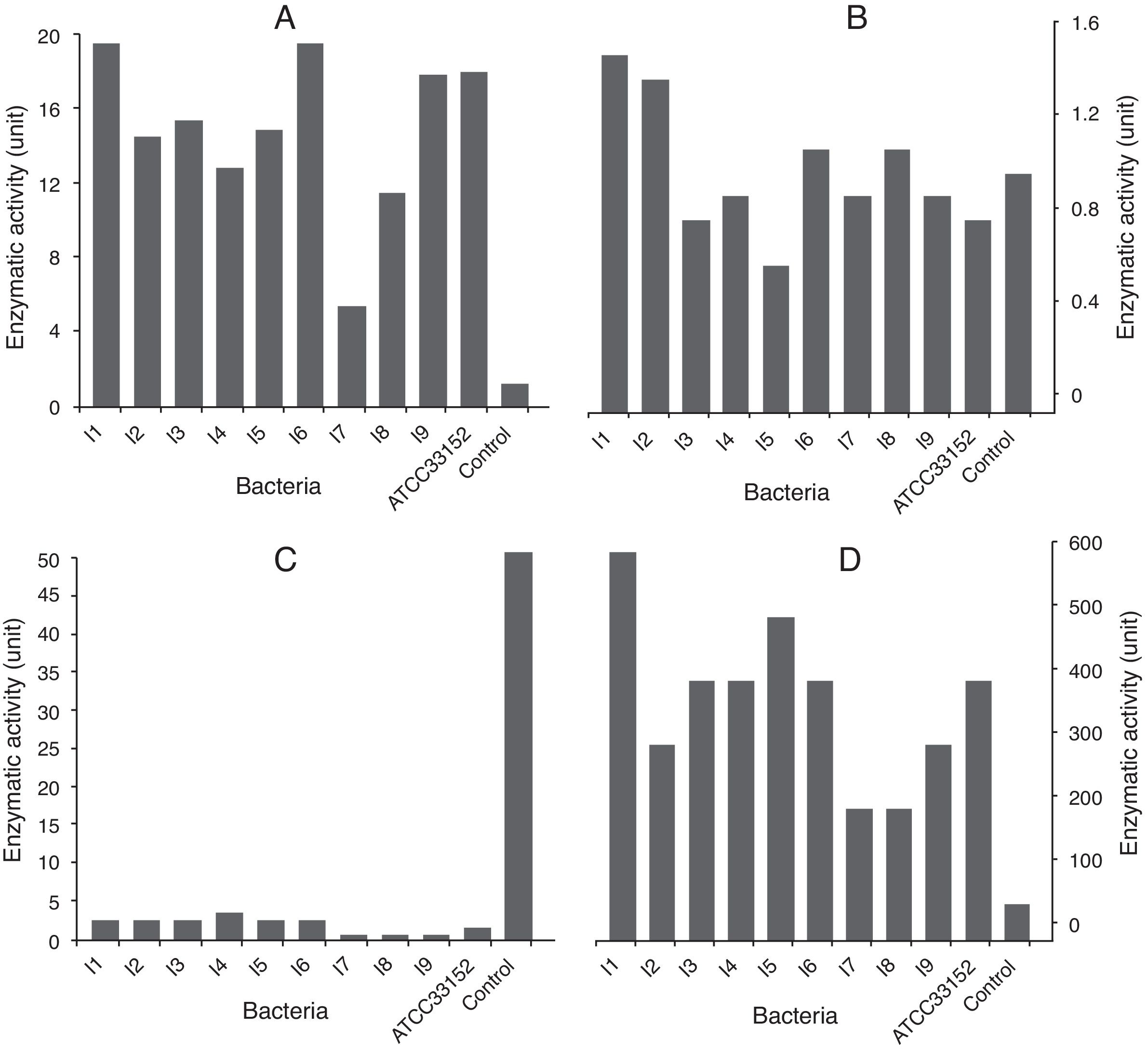
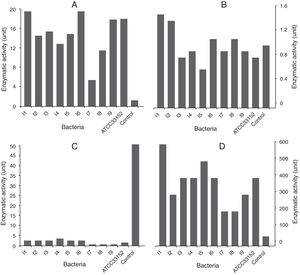
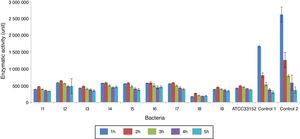
The isolates 1 and 6 had higher PLA activity. The cell lysates of these strains exhibited meaningful statistical differences from those of other strains (p<0.05). The weakest PLA activity was shown by the isolate 7. When the culture supernatants were studied, only the isolates 1, 2 and 8 were found to be capable of secreting PLA (Fig. 1A and B). Nevertheless, a statistically meaningful difference was not found between isolates 1 and 2 (p>0.05), while there was a statistically significant difference between each of those isolates and the isolate 8 (p<0.05).
The highest PLC activities in the lysates and supernatants were produced by the lysates of the isolate 4 (3U), and the supernatant of the isolate 1 (600U). Moreover, all the cell lysates demonstrated weak PLC activity (1, 2 and 3 units were produced by the cell lysates of ATCC 331452; the isolates 1, 2, 3, 5 and 6, and the isolate 4, respectively), except for those of the isolates 7, 8 and 9 (Fig. 1C and D).
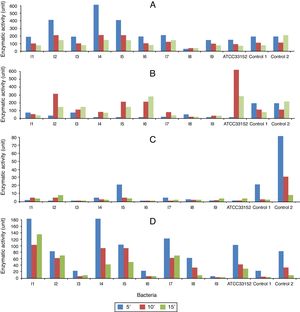
The alkaline phosphatase activities of the L. pneumophila lysates decreased with time. In all the measurements, the highest and lowest activities were produced by the isolates 2 and 8, respectively. Extracellular enzyme activity was only found in cultures of the isolate 4 (1U), starting from the second hour of cultivation (Fig. 2). Acid phosphatase activities of L. pneumophila in the lysates and supernatants were not determined.
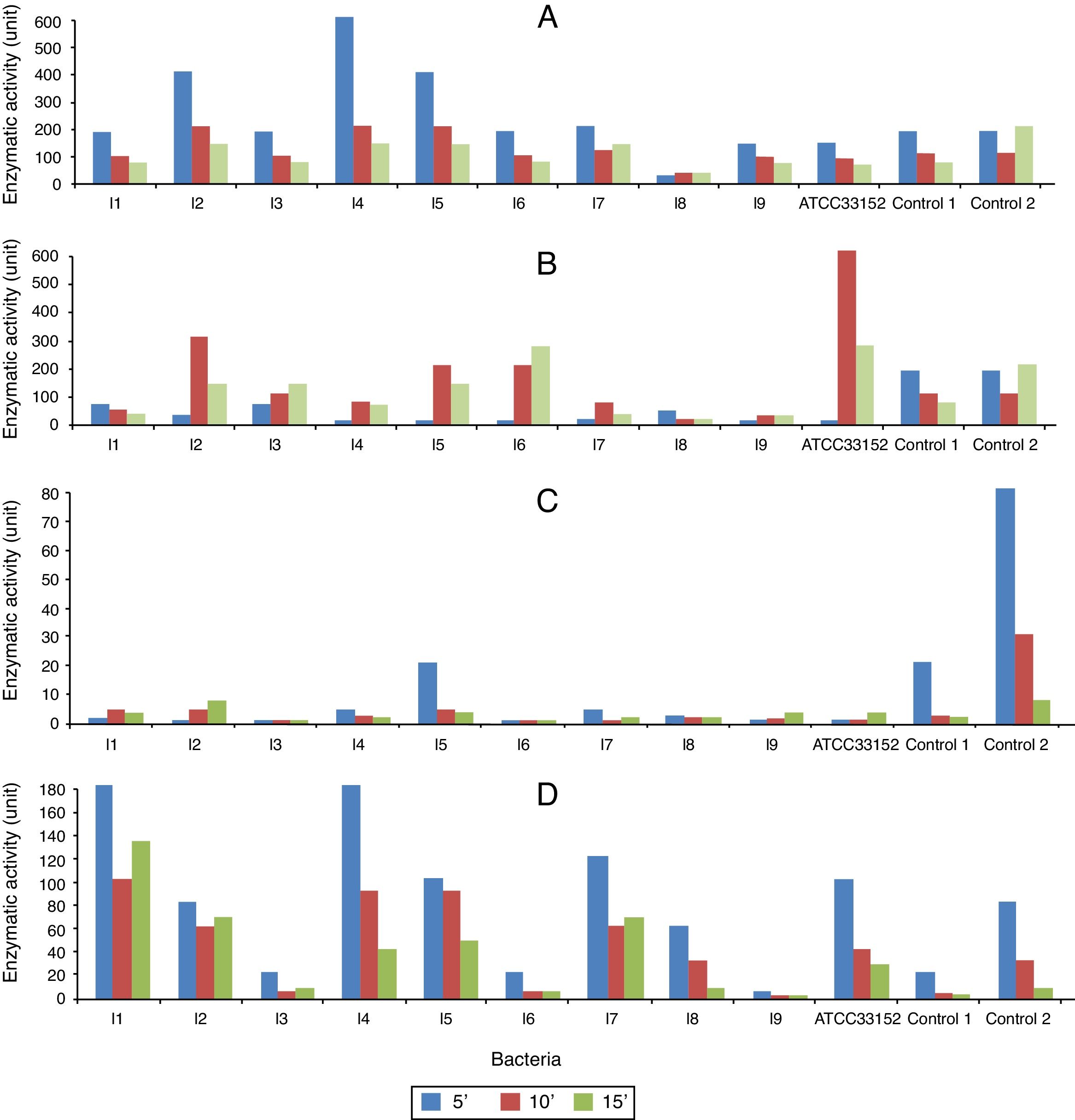
In the assays measuring pNP formation from p-NPC, the highest lipase activity was found in the cell lysates of the isolate 4 (600U) (Fig. 3A). Of all the isolates, the extracellular lipase level was highest in the cultures of isolate 6 (300U); in comparison, the activity measured in the control cultures was 600U (Fig. 3B).
The lipase activity of the cell lysates measured using p-NPP as a substrate was weaker than that measured using p-NPC as a substrate. In the first 5min, the isolate 5 showed the highest lipase activity (20U), while the isolates 3 and 6 did not exhibit any activity (Fig. 3C and D). The extracellular L. pneumophila lipase activities (using p-NPP as substrate) were more pronounced than those of the cell lysates. The highest lipase activities were detected in the isolates 1 and 4 (180U) and in the isolate 7 (120U).
Any intracellular or extracellular resorufin esterase activity was not found in any of the cultures. Moreover, the cell lysates of L. pneumophila cultures were not capable of forming free fatty acids from 1-MG. Only the supernatant of the isolate 5 was able to degrade 1-MG, and none of the lysates or supernatants contained any enzymatic activity capable of converting 1,2-DG to free fatty acid.
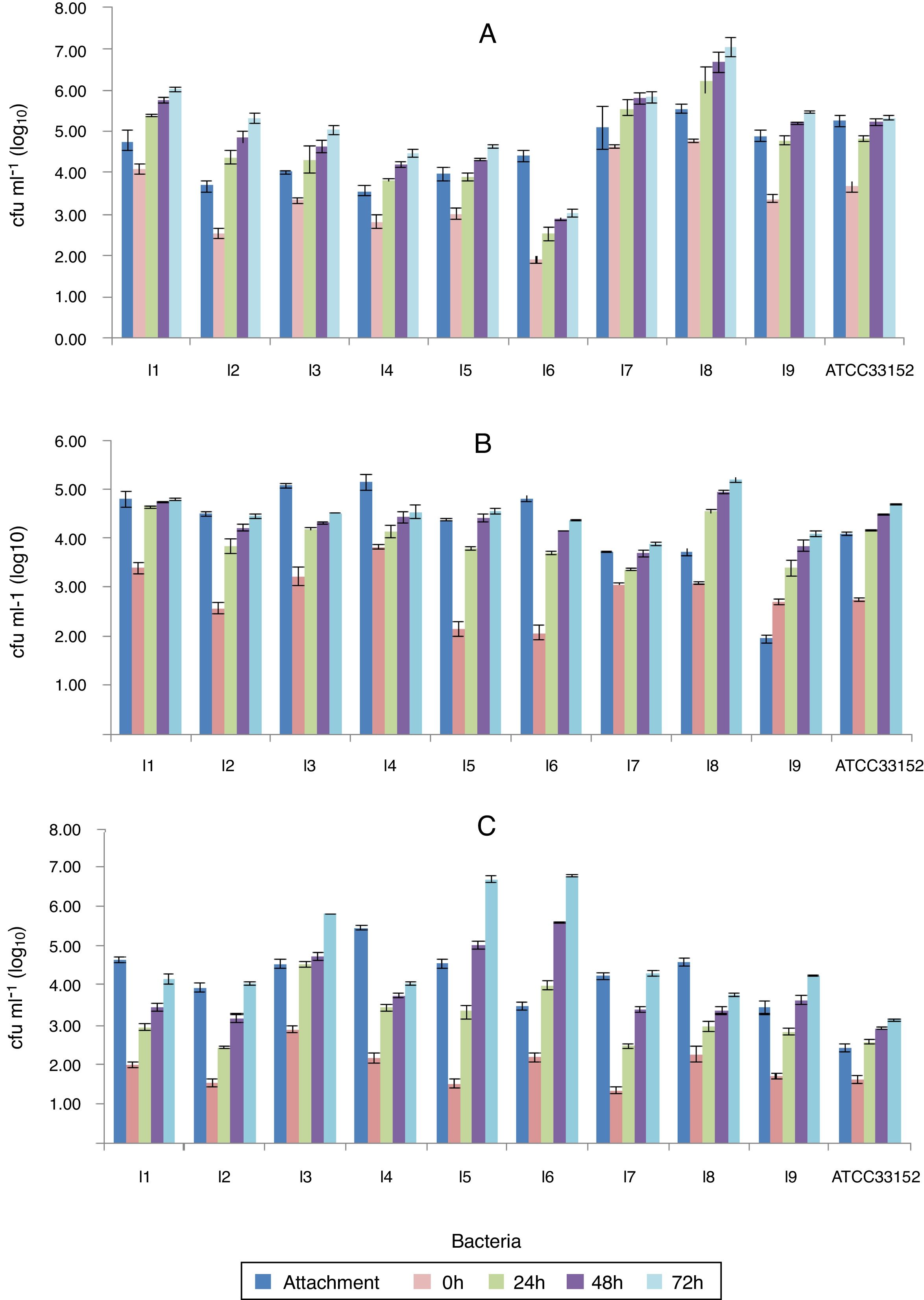
The investigation of the interaction between L. pneumophila and its hostsThe highest ability to adhere to PBMCs was found in the isolate 8 (63%), while the lowest was found in isolate 4 (43%) (the percentages reflect adherence relative to the positive control). The isolate 7 penetrated and reproduced in PBMCs most efficiently (ninety-one percent of the bacteria penetrated).
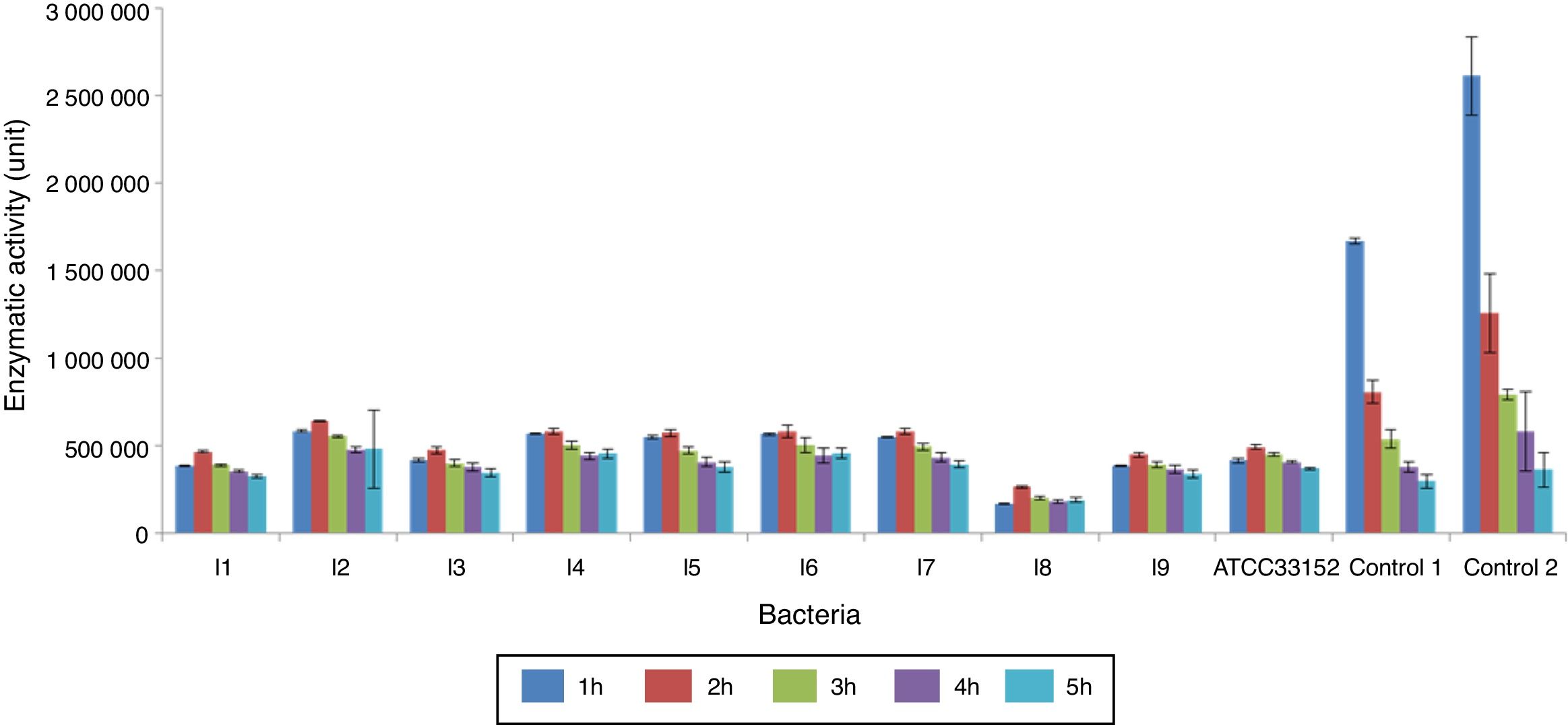
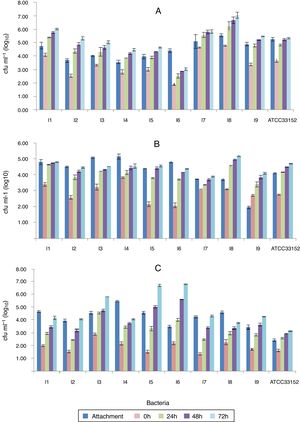
The bacteria continued to divide over a period of three days and the fastest growth rate was observed in the first 24h (Fig. 4). Between all the time points, a statistically meaningful difference was not found with respect to the intracellular growth (p<0.05), except isolate 6. When the number of bacteria that can penetrate the host cells was compared with the growth rate inside the host, the isolate 2 was the most successful strain. It exhibited 1.83-log, 2.02-log and 2.16-log increases at the end of 24, 48, and 72h, respectively (Fig. 5A).
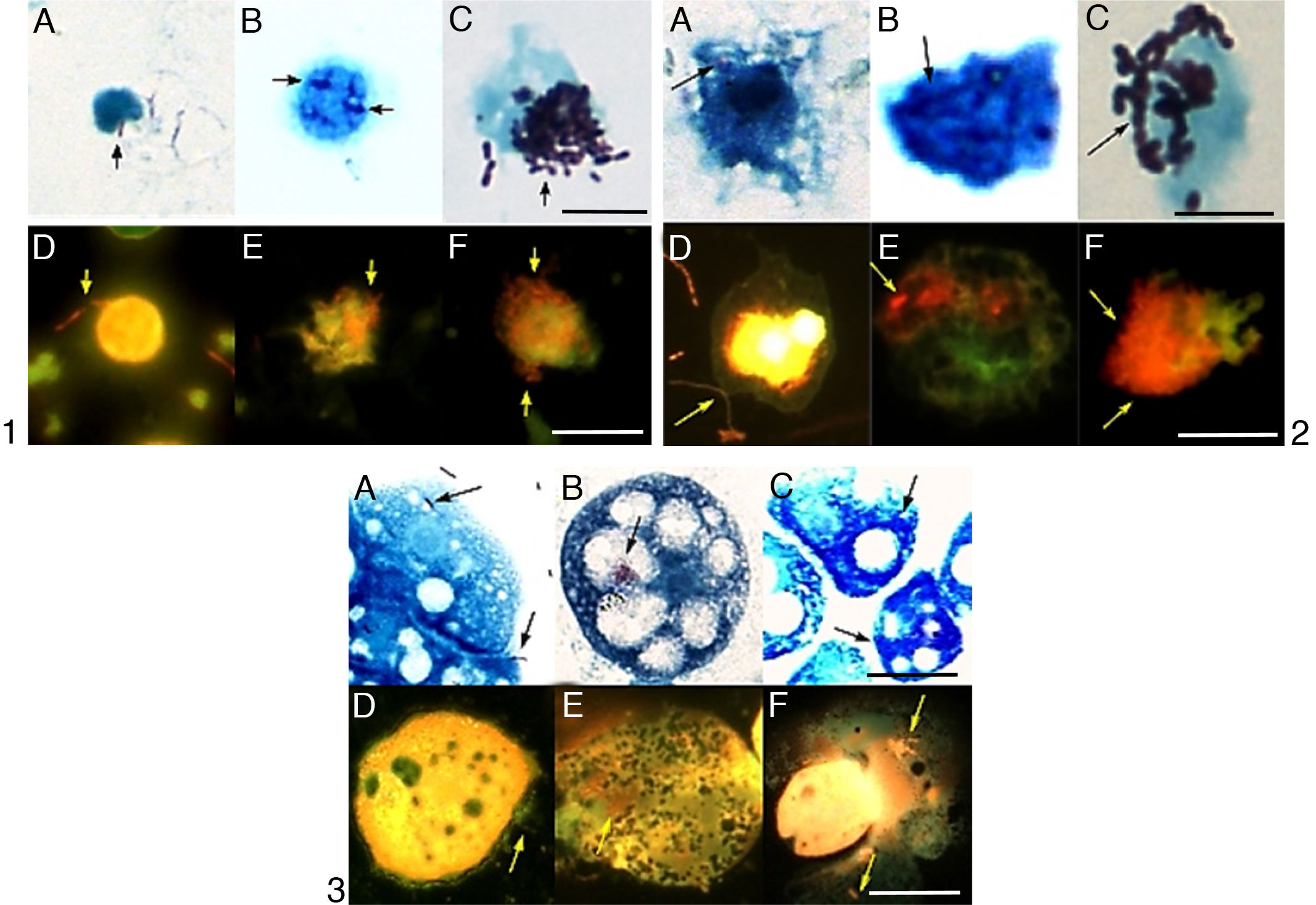
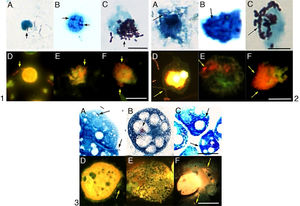
The intracellular growth of bacteria in different host cell. 1: the I2 in PBMCs; 2: the I5 in PMs; 3: the I5 in A. castellanii. A, B, C: penetration, intracellular growth at the 24th and 72nd hours in light microscopy; D, E, F: penetration, intracellular growth at the 24th and 72nd hours in epifluorescence microscopy. Sacale Bar=10μm in Picture 1; 25μm in Picture 2; 50μm in Picture 3 (100×).
The isolates that exhibited the highest and lowest adhesion to PMs were isolate 3 (54%) and isolate 9 (22%). The isolate 9 showed the highest host-penetration (139% relative to the control). The bacteria continued to divide inside the PMs for 3 days; however, the fastest growth was observed in the first 24h. When the number of bacteria growing in the PMs was compared to the number of the penetrating bacteria, the fastest reproduction rates were observed in the isolate 5, which showed 1.64-log, 2.26-log and 2.4-log increases by 24, 48 and 72h, respectively (Figs. 4 and 5B).
Of all L. pneumophila isolates, the isolate 4 exhibited the highest adhesion (63%) to A. castellanii trophozoites. There was a statistically meaningful difference (p<0.05) between the isolates with respect to their ability to attach to the trophozoites. The control strain ATCC 33152 had the highest ratio of penetration into A. castellanii trophozoites (67%). This bacterium was grown in the trophozoites for 3 days, and the fastest growth was observed in the first 24h. Of all the isolates, the isolate 5 grew most efficiently in the trophozoites; i.e., it exhibited 1.80-log, 3.49-log and 5.17-log increases by 24, 48 and 72h, respectively (Figs. 4 and 5C).
DiscussionUpon the attachment of microorganisms on a surface and the secretion of polysaccharide products, a bacterial biofilm is formed. Biofilm helps bacteria obtain nutrients and protects them from excess temperatures and pH, chemicals and predators.25L. pneumophila is known to form biofilms in nature, and there is a study demonstrating that they form monospecies biofilms in artificial media as well.7 The study serves us to better understand how bacteria adapt from growing as free cells to growing in biofilms and the effect of this conversion on the organism. Amoeba cells are natural hosts of Legionella and are different in the sense that they utilize phagocytosis for feeding and not for defense. Thus the attachment and penetration of Legionella to amoeba is different than the attachment and penetration involved in the mammalian phagocytosis of cells.2,26 Nevertheless, these bacteria form biofilms to survive in water, and they use amoeba species as hosts to reproduce.27 This could explain the correlation between the adhesion of the isolate 4 to inorganic surfaces and its adhesion to A. castellanii trophozoites. When 14-day growth of the isolates in biofilms were compared with their growth in PBMCs, PMs or amoeba trophozoites, a meaningful and strong positive correlation was found only between the growth in biofilm and the growth in trophozoites (r=0.72, p<0.05). This indicates that, similar to the findings regarding adhesion to inorganic surfaces or host cells, bacterial growth properties also vary depending on the habitat.
Flagella help L. pneumophila in motility, finding the host, biofilm colonization, and virulence.28,29 All of the isolates studied here were mobile and had polar flagella. Therefore, it could be postulated that these were virulent strains.
Pili are organelles responsible for bacterial attachment to host mucosa. Stone and Abu Kwaik30 showed two different types of pili on L. pneumophila surfaces. One of these types of pili was the type IV pilus, which does not require complementation to attach to mammalian host cells. The result about hemagglutination assays emphasizes the importance of hemagglutination activity, and thus, the presence of the type IV fimbria as a virulence factor, in providing information on bacterial pathogenicity.
The hemolytic activity of L. pneumophila was first shown on the mammalian blood cells in Feeley Gorman Agar (FG agar) plates.31 In the current study, the hemolytic enzyme secretion continued until day 6, and the hemolytic activity was more pronounced on human erythrocytes than in sheep erythrocytes. This could be caused by differences in the media compositions and erythrocyte suspensions used. In some of the eBYEA plates, semi-transparent and greenish α-hemolysis zones (wide-zone β-hemolysis) were also detected around the β-hemolysis zones. The zone diameters in these cases were measured as the diameter of the α-hemolysis zone. Therefore, the α-hemolysis zone diameters were larger when both α- and β-hemolysis was present. It was previously shown that bacteria such as human pathogen streptococci, Corynebacterium pseudotuberculosis and Staphylococcus aureus, and yeast such as Candida albicans produce wide-zone β-hemolysis.32 To our knowledge, Legionella species were not previously reported to cause this kind of hemolysis. In fact, different proteins are responsible for different types of hemolysis in bacteria.33 This rule could apply to our isolates as well, and different enzymes are selectively effective on human or sheep erythrocytes. In recent studies, hemolytic index calculations were employed in hemolysis studies.34 In the current study, a meaningful correlation (r=0.761–0.996; p<0.05) between the hemolysis zone diameters and the hemolytic index values was detected.
Legionella bacteria produce proteases. These enzymes may play roles in hemolysis and are considered weak virulence factors. A meaningful correlation between protease activity and hemolytic activity (based on hemolytic indexes calculated using human and sheep erythrocytes) was not found (p>0.05). Our finding that trypsin and chymotrypsin were not produced in the cultures is consistent with previously reported results with Legionella.35 Although it was the most virulent strain, the isolate 2 had weaker proteolytic activity than the other isolates. This result shows that the relationship between proteolytic activity and virulence is not compelling. A meaningful negative correlation was found between protease activity and growth in PBMCs (r=−0.634; p<0.05). This is further evidence demonstrating that virulence factors vary with the bacteria and host species.
The type II secretion system-controlled nuclease production by the isolates was also investigated in the current study. Only isolate 1 produced higher DNase and RNase activity compared to ATCC strain 33152. In correlation analysis to determine the association between DNase and RNase activities produced by the isolates, a meaningful positive correlation was detected (r=0.834; p<0.01). The isolate 8 had both the lowest RNase activity and growth capability in trophozoites, supporting the statements by Rossier and Cianciotto36 that there is a connection between nuclease activity and factors that play roles in bacteria-host relationships. On the other hand, Gunderson et al.37 remarked that nuclease activity of L. pneumophila has a critical importance for promoting infection of amoebae.
Phospholipases constitute an important subset of bacterial virulence factors. Cellular disintegration caused by phospholipid degradation by bacterial phospholipases is one of the fundamental steps in bacterial infections. Very limited secretion of PLA was found in L. pneumophila cultures. This result supports the statement by Baine38 that PLA is a cell wall enzyme. Nevertheless, another study opposes these findings, i.e., extracellular the PLA activity could be higher.11 This conflicting result could be due to the use of bacteria in different growth phases, and PLA secretion could replace PLC.15 In the current study, a meaningful positive correlation was found between PLA activity in the cell lysates and the PLC activity in the supernatants (r=0.648; p<0.05). Hence, it seems that similar enzymes are responsible for PLA and PLC activities. This conclusion is also supported by our results that the isolate 1 demonstrated the highest PLA and PLC activities in the supernatants. When the relationship between the PLA/PLC activities and pathogenicity against the host was investigated, a meaningful positive correlation was also observed between the attachment to PMs and the PLC activity (r=0.818; p<0.01).
Phosphatases constitute another family of the type II secretion system enzymes with significance in host invasion and virulence in the genus Legionella. Cell-bound alkaline phosphatases and extracellular acid phosphatases were previously reported in this genus.11,39 Alkaline phosphatases work in the periplasmic space and enable bacteria to utilize inorganic phosphate.11,15 In the current work, the L. pneumophila cell lysates showed the alkaline phosphatase activity as verified by the API ZYM test system. This activity decreased after 2h of incubation, indicating that substrate levels decreased in a time-dependent manner. Moreover, a meaningful positive correlation was observed among the alkaline phosphatase activities measured at five different time points (r=0.953–0.976; p<0.01). Although the experiments measuring acid phosphatase activity were duplicated with two different p-nitrophenol phosphate substrates (N1891 and 73737, Sigma–Aldrich Chemical Co., Inc.) no acid phosphatase activity was detected in L. pneumophila supernatants and lysates, unlike in other studies. However, in the API ZYM test system, the acid phosphatase activity was detected in all the isolates. The isolates 5, 6 and 8 showed the strongest positive results.
The genus Legionella was first reported to possess lipase activity by Baine et al.31 Aragon and his colleagues11 defined the activity as monoacyl esterase lipase activity. The enzyme is regulated by the pilD gene and plays a role in tissue decomposition in the host. High p-NPC activity was earlier reported in the supernatants of a standard Philadelphia-1 strain 130b. They also showed that the bacteria could use this lipase both intracellularly and extracellularly. In our study, the cell lysates of the isolate 5, which had the strongest p-NPP activity, possesses the highest growth rate in PMs and trophozoites, so there could be a relationship between lipase activity and growth of this strain in the host. When the connection between L. pneumophila p-NPC/p-NPP activities and pathogenicity toward the host cell was investigated, a meaningful correlation was observed between the p-NPP activity of the L. pneumophila cell lysates and pathogenicity in amoeba trophozoites (p<0.05). This finding supports the idea that L. pneumophila lipase activity and pathogenicity are related. Aragon et al.40 also demonstrated that PLC and lipase activities are correlated with and important in host pathogenicity. A meaningful positive correlation was observed between the lipase activity of cell lysates (using p-NPC substrate) and PLC activity (p<0.05).
The current intracellular growth assays have validated the results of previous studies.41 Additionally, the growing in trophozoites indicates that among all three hosts, A. castellanii trophozoites induce bacterial growth most efficiently. Thus, adhesion and penetration of the host cell as well as growth capability in the host vary within bacterial strains and the host cell type. It could be concluded that adhesion, penetration and growth are regulated by different factors. The results about bacterial infectivity of host cells of the isolate 6 show that L. pneumophila exhibits different levels of pathogenicity in different hosts. In fact, It has been reported that L. pneumophila use different mechanisms for infecting amoebas.42
When the results of pathogenicity experiments with L. pneumophila on different hosts were compared, the differences observed from host to host became even more pronounced. There was a meaningful correlation between bacterial adhesion to PBMCs and adhesion to PMs (r=0.738; p<0.01). Another meaningful correlation was detected between penetration into PBMCs and attachment to A. castellanii trophozoites (r=0.649; p<0.05). Moreover, it was found that the level of pathogenicity in the host varies with the type of pili and surface proteins as well as the enzymes produced by the bacteria.43 These results are consistent with ours and confirm the conclusion that L. pneumophila exhibits different mechanisms of pathogenicity according to the host type.
When the virulence factors were analyzed, every isolate appeared to have different virulence capabilities. However, considering all the results, the isolate 4 was the most effective bacteria in 34% of the experiments, followed by the isolate 2 (22%), 1 (17%), 5 (13%), 3 and 7 (5%), 6 and ATCC 33152 (4%), 8 (3%), and 9 (1%). At the same time, it was found that the isolate 4 was more effective than standard bacteria in 73% of the experiments, following by the isolates 1 (66%), 6 (64%), 5 (61%), 2 (59%), 3 and 7 (57%), 9 (42%), and 8 (40%).
On the other hand, when the results were evaluated in terms of the type II and type IV secretion systems, isolate 4 was the most virulent bacterial strain according to the type II secretion system, while isolate 2 was the most virulent bacterial strain according to the type IV secretion system.
The growing media is very important for bacterial pathogenicity20 and this was indicated by the fact that the infection capacity of L. pneumophila decreased after growing on BCYE medium. In the future studies, we plan to investigate the infection capacity of our isolates which may change after they growth in amoeba cultures. On the other hand, for the detection of virulence of these bacteria, they should also be investigated when left in nature. Therefore, we plan to study the growth of Legionella in a cooling tower system model.
Conflicts of interestThe authors declare no conflicts of interest.
This work was supported by the Research Fund of Istanbul University (Project number: 2747).