Bacteroides fragilis is the strict anaerobic bacteria most commonly found in human infections, and has a high mortality rate. Among other virulence factors, the remarkable ability to acquire resistance to a variety of antimicrobial agents and to tolerate nanomolar concentrations of oxygen explains in part their success in causing infection and colonizing the mucosa. Much attention has been given to genes related to multiple drug resistance derived from plasmids, integrons or transposon, but such genes are also detected in chromosomal systems, like the mar (multiple antibiotic resistance) locus, that confer resistance to a range of drugs. Regulators like MarR, that control expression of the locus mar, also regulate resistance to organic solvents, disinfectants and oxygen reactive species are important players in these events. Strains derived from the parental strain 638R, with mutations in the genes hereby known as marRI (BF638R_3159) and marRII (BF638R_3706) were constructed by gene disruption using a suicide plasmid. Phenotypic response of the mutant strains to hydrogen peroxide, cell survival assay against exposure to oxygen, biofilm formation, resistance to bile salts and resistance to antibiotics was evaluated. The results showed that the mutant strains exhibit statistically significant differences in their response to oxygen stress, but no changes were observed in survival when exposed to bile salts. Biofilm formation was not affected by either gene disruption. Both mutant strains however, became more sensitive to multiple antimicrobial drugs tested. This indicates that as observed in other bacterial species, MarR are an important resistance mechanism in B. fragilis.
Bacteroides fragilis, an anaerobic Gram negative bacterium that colonizes the gastrointestinal tract of humans, is a minor component of the human fecal microbiota.1 Despite its beneficial role as a member of the gut microbiota, B. fragilis is the strict anaerobic bacteria most commonly isolated from human infection.2 This duality in the interaction with the host is well recognized and several virulence factors have been described to account for this behavior, including a capsular polysaccharide complex (CPC),3 adhesins and the Bacteroides fragilis toxin (BFT).4 The high tolerance to oxidative stress (OS) observed in this species is regarded as a major advantage when invading oxygenated host tissues, such as the peritoneal cavity5 and resisting the oxidative burst imposed by macrophages.6 An intense modulation of gene expression takes place in B. fragilis during oxidative stress as shown by microarray studies,7 where the expression of over 45% of the total genome of the bacteria is affected. Several OS detoxifying enzymes are found in this species, such as catalase (KatB), superoxide dismutase (SOD), Dps, thioredoxins and alkyl hydroperoxide reductase (AhpC), but the regulation of the OS response in B. fragilis is complex and not fully understood. The transcriptional regulator OxyR has received attention in the past decades and is recognized as an important player during OS.8 Studies with OxyR mutants in B. fragilis however, have shown that despite its importance, OxyR is not the sole mediator of the OS response and other regulators are involved.9
Recently, we described the role of a transcriptional regulator homolog to the multiple antibiotic resistance regulator (MarR) family in the OS response of B. fragilis.10 Members of the MarR family of transcriptional regulators have been described as winged helix proteins that bind directly to the DNA to control a wide range of biological processes in both bacteria and archaea.11,12 So far, the best known model of regulation by MarR family members is the regulation of multiple antibiotic resistance in Escherichia coli by the prototypical member MarR, described by George and Levy in 1983.13 Since then, several members of this family have been characterized as having an important role in microbial pathogenesis and resistance to antimicrobials although the mechanism in which the regulation occurs may change in different species.14 Transcriptional regulators of the MarR family are known to affect both the oxygen stress response and the expression of virulence factors in Gram negative pathogens such as Salmonella enterica and E. coli.15 According to annotations in the genebank database (NCBI) some strains of B. fragilis may carry up to four different homologs of genes coding for MarR proteins in its genome, but their role in the physiology of this species has not been studied in detail. Multidrug resistance in bacteria is regarded as a major challenge for clinicians in the world. Strict anaerobic bacteria are particularly challenging, since most hospitals and clinical settings, particularly in developing countries, lack of facilities and equipment used for cultivation under strict anaerobiosis. Research labs have shown that multidrug resistance (MDR) in Bacteroides is on the rise for the past few decades.16–18 Resistance to most classes of antimicrobial drugs used in clinical settings was already reported for Bacteroides species, including carbapenems and metronidazole.18,19 Production of broad spectrum β-lactamases in B. fragilis is well documented, and at least three enzymes coded by the genes cepA, cfiA and cfxA were already described,18 but the mechanisms regulating the expression of resistance determinants in anaerobes are poorly understood. To address such issues, in the present study we disrupted the expression of two genes coding for proteins of the MarR family in B. fragilis and investigated the effect in antimicrobial resistance, susceptibility to oxidative stress and biliary salts, and formation of biofilms in vitro. As previously observed, interfering with transcriptional regulators of the marR family (Teixeira 2013), leads to altered phenotypes and response to environmental stress in B. fragilis.
MethodsBacterial strains and plasmidsBacterial strains and plasmids used in this study are listed in Table 1. The B. fragilis strains were routinely grown in brain and heart infusion agar supplemented with menadione (10μg/mL) and hemin (0.5μg/mL) (BHI) and in pre-reduced and anaerobically sterilized BHI broth (BHI-PRAS). Cultures were grown at 37°C in an anaerobic chamber (Coylabs, Michigan, USA) with an atmosphere of 80% N2, 10% CO2 and 10% H2. The antibiotics rifampicin (25μg/mL), erythromycin (10μg/mL) and gentamycin (100μg/mL) were added to the media when required. The E. coli strains were routinely grown in lysogeny broth (LB), containing 1% triptone (w/v, Difco, BD, New Jersey, USA), 0.5% yeast extract (w/v) (Oxoid, Thermo-Fisher, Massachusetts, USA) and 0.5% NaCl (w/v) or LB agar (1.5% agar-agar, w/v, Difco). Ampicillin (100μg/mL), tetracyclin (10μg/mL), spectinomycin (50μg/mL) and kanamycin (50μg/mL) were added to the medium when required. For cloning and electroporation, super optimal broth with catabolite repression (SOC) medium containing 2% triptone, 0.5% yeast extract, 0.05% NaCl, 2.5mM KCl, 10mM MgSO4·10 MgCl2 and 20mM glucose, pH 7.0, was used. Antibiotics were purchased from Sigma–Aldrich, Missouri, USA.
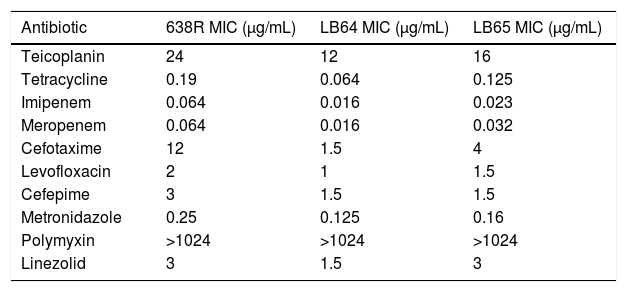
E-test results of in vitro activity of ten antibiotics against wild type B. fragilis strain 638R and mutant strains LB64 and LB65.
| Antibiotic | 638R MIC (μg/mL) | LB64 MIC (μg/mL) | LB65 MIC (μg/mL) |
|---|---|---|---|
| Teicoplanin | 24 | 12 | 16 |
| Tetracycline | 0.19 | 0.064 | 0.125 |
| Imipenem | 0.064 | 0.016 | 0.023 |
| Meropenem | 0.064 | 0.016 | 0.032 |
| Cefotaxime | 12 | 1.5 | 4 |
| Levofloxacin | 2 | 1 | 1.5 |
| Cefepime | 3 | 1.5 | 1.5 |
| Metronidazole | 0.25 | 0.125 | 0.16 |
| Polymyxin | >1024 | >1024 | >1024 |
| Linezolid | 3 | 1.5 | 3 |
B. fragilis 638R mutants were generated by insertional inactivation using the suicide plasmid pFD516.20 Briefly, a fragment of 300bp of the gene BF638R_3159 (For-5′GGATGGAATTCCGCCAGGCCGTTC and Rev-5′CATAGACGTCGTTGATCAGGGGTTTCACG) and 252bp of the gene BF638R_3706 (For-5′ATCGGAATCCATACCGGAACTTCAGACAG and Rev-5′TTCCTGCAGCTCTTTTCCTGTTCTGG) were amplified by polymerase chain reaction (PCR) and cloned into the cloning vector pGEM®-T Easy (Promega). The sequences of the genes were obtained from Genbank under the accession number FQ312004.1. The PstI/EcoRI fragments, generated after digestion of the cloning vector, was cloned into the PstI/EcoRI sites of the suicide plasmid pFD516 and transformed in electrocompetent E. coli DH10b strains. The resulting plasmid was mobilized into B. fragilis 638R by triparental mating.21 Transconjugants were selected on BHI containing rifampicin (20μg/mL), erythromycin (2μg/mL) and gentamicin (100μg/mL). Insertion of the plasmid in the correct loci was confirmed by PCR using M13 primers (IDT, Iowa, USA) as described above.
Oxygen stress survivalTo prepare the inoculum, bacterial strains were grown for 48h at 37°C in BHI-PRAS and the cells were harvested by centrifugation at 16,000×g for 5min. Cells were resuspended in phosphate buffered saline solution (PBS) pH 7.3 to an OD600=1. An inoculum of 10μL of decimal serial dilutions up to 10−7 were spotted in BHI plates containing rifampicin (25μg/mL) and gentamycin (100μg/mL) and then incubated at 37°C in aerobiosis. Culture plates were transferred to the anaerobic chamber at each time point of 24, 48 and 72h and further incubated for 48h in anaerobiosis. Plates not exposed to oxygen were used as controls. The results were scored as the lowest dilution where growth could be observed in each time point.
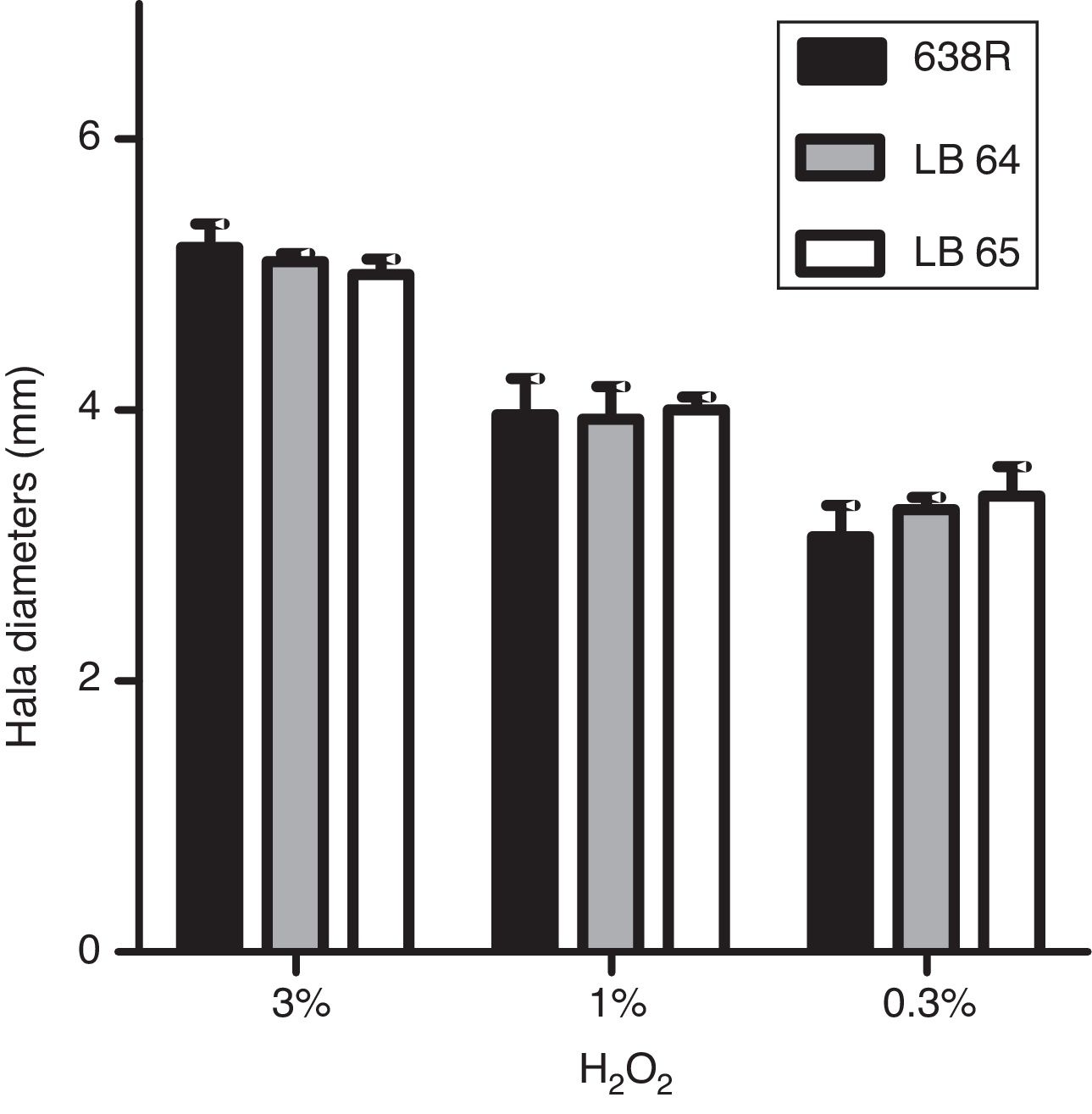
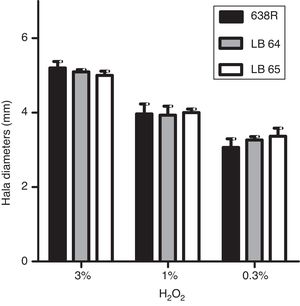
Hydrogen peroxide susceptibilityBacterial strains were initially grown for 48h at 37°C in BHI-PRAS adjusted to the McFarland scale 1 (3×108UFC/mL) and spread with a swab in Brucella agar plates to achieve confluent growth. Sterile filter paper disks (6mm, Laborclin) containing 10μL of hydrogen peroxide solutions (0.3%, 1% and 3%) were placed on the agar surface and the plates were incubated for 48h at 37°C in anaerobiosis. Diameters of the growth inhibition zone were measured in millimeters.22 All experiments were performed in triplicates and statistical analysis was performed as described above (ANOVA).
Susceptibility to biliary saltsThe minimal inhibitory concentration (MIC) and minimal bactericidal concentration (MBC) of biliary salts were performed as described before.23 To prepare the inoculum the strains were grown in BHI-PRAS supplemented with antibiotics for 18h at 37°C and then inoculated in fresh BHI-PRAS until an OD600 of 0.4 was reached. Bacterial growth was diluted 1:10 and 20μL were inoculated in wells of a 96-well plate containing 180μL of BHI-PRAS with biliary salts dilutions ranging from 0 to 20%. Plates were incubated for 48h at 37°C and growth was measured by Reading the OD600 in a plate reader (Multiskan, Molecular Devices, USA). The MIC was determined as the last concentration where the OD reading was superior to the blank well (non-inoculated well). For the MBC, three concentrations above the MIC were plated in BHI agar with antibiotics and incubated for 48h at 37°C. Growth was evaluated by visual inspection. Three individual experiments conducted in triplicates were performed.
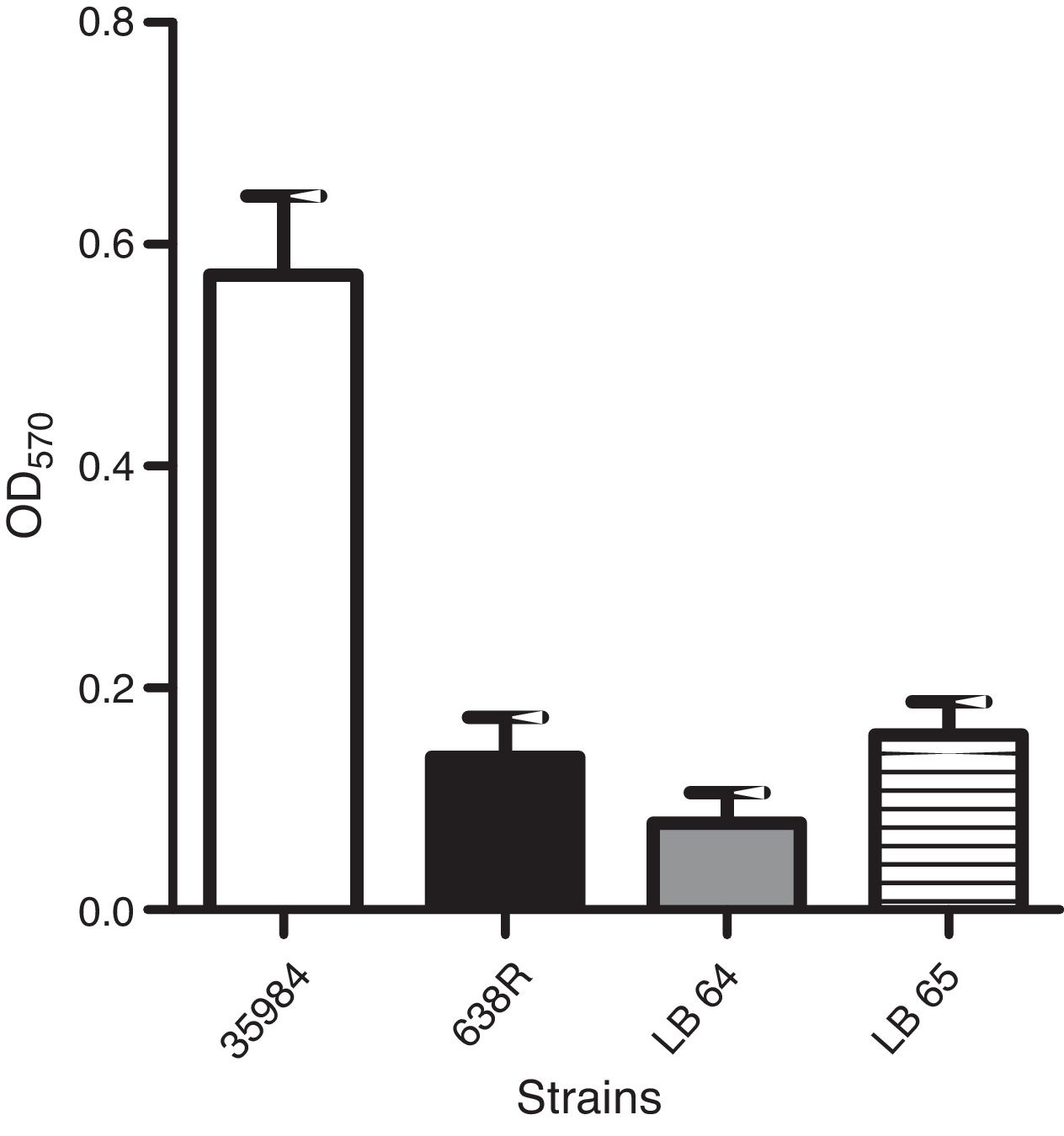
Biofilm formationAssays for biofilm formation were done specifically for anaerobes as described in the literature.24 Bacterial strains were inoculated in 96-well plates in PRAS – BHI supplemented with hemin (0.5mg/mL – Sigma–Aldrich, USA) and 1% glucose (w/v) and incubated for 24h at 37°C in an anaerobic chamber. The media was then removed and carefully washed with 200μL of phosphate buffered saline solution (PBS – 10mM sodium phosphate, 150mM NaCl, pH 7.4). The wells were dried for 1h at 60°C and stained with 2% Gram's violet crystal. Excess dye was washed with distilled water and dried for 10min at 60°C. Intracellular dye was solubilized with 150μL of 33% glacial acetic acid in water (v/v) and the optical density at an OD570nm was measured for each well. All experiments were conducted in triplicates. Non-inoculated wells were used as background controls. Statistical analysis was performed as described above (ANOVA).
E-testAntimicrobial susceptibility tests were performed using E-tests strips according to the manufacturer's instructions (Biomerieux, France) and the CLSI guidelines (Clinical Laboratories Standards Institute). Bacterial strains were initially grown for 48h at 37°C in BHI-PRAS and adjusted to the McFarland scale 1 (3×108UFC/mL) and spread with a swab in Brucella agar plates supplemented with 5% defibrinated sheep blood to achieved confluent growth. E-test strips were placed in the plates with the help of sterile forceps and the plates were incubated in anaerobiosis for 48h at 37°C.
ResultsSurvival after exposure to atmospheric oxygen is reduced in MarRI and MarII mutantsOur previous results show that the inactivation of a MarR homolog (BmoR) in a strain of B. fragilis leads to increased sensitivity to oxygen.10 The expression of two genes (BF638R_3159 and BF638R_3706) coding proteins with homology to transcriptional regulators of the MarR family was interrupted by insertion of a suicide plasmid in their coding sequence. Strains carrying the plasmid inserted into the locus BF638R_3159 and BF638R_3706 are respectively referred as LB64 and LB 65. To test if the single inactivation of the two remaining MarR homologs in B. fragilis has a similar effect, 10-fold serial dilutions of the bacterial strains were spotted on agar plates and exposed to atmospheric oxygen for up to 72h. Bacterial susceptibility was evaluated by growth recovery after exposure to oxygen in increasing time points. As anticipated the wild type strain 638R and both mutant strains, LB64 and LB65 presented confluent growth when grown in strict anaerobiosis, even at the maximum dilution (10−7). Difference among the strains was noted as soon as after 24 and 48h of exposure to oxygen. Growth was reduced by 2 orders of magnitude for both mutant strains (10−2) when compared to the wild type strain (10−4). After 72h of exposure growth was only observed in the wild type strain 638R. This result indicates that the absence of either of the two MarR homologs affect resistance to OS.
Susceptibility to hydrogen peroxide is not affected by inactivation of MarR homologsTo confirm the role of MarR homologs in the OS response, susceptibility to hydrogen peroxide was evaluated by disk diffusion in Brucella agar plates. The diameters of inhibition halos were compared between the parental and mutant strains. All strains were susceptible to the three concentrations of H2O2 tested (0.3%, 1% and 3%) and no significant changes were observed in growth of the mutant strains (Fig. 1).
Inhibition of bacterial growth by disk diffusion of hydrogen peroxide in solid media. Growth of wild type strain 638R of B. fragilis was compared to strains LB64 (BF638R_3159) and LB65 (BF638R_3706) carrying mutations in gene homologs to marR. Inhibition halo in diameters is shown with standard deviations.
Resistance to biliary salts is important for intestinal bacterial residents. Mechanisms of resistance include alterations of the permeability of the external membrane and efflux pumps.25 Since MarR is known to regulate the expression of outer membrane proteins, we evaluated whether interrupting genes homologs to marR would compromise survival in high bile concentrations. After 48h of incubation, all the strains were inhibited by concentrations above 10% of biliary salts. Three concentrations above the MIC were plated to determine the MBC. Again, all three strains were killed by 12% of biliary salts indicating that the MIC and MBC was identical among the three strains tested.
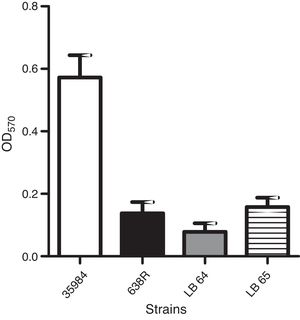
Production of biofilm in polystyrene surfaces was observed in the wild type strain as previously described.24 A strain of Staphylococcus epidermidis (ATCC 35984) was also used as a positive control for the assay. Capacity to form biofilm in vitro was not affected in the mutant strains when compared to the wild type (Fig. 2).
Antimicrobial susceptibility evaluated by E-testAntibiotic susceptibility testing was performed using E-test strips in accordance to CLSI guidelines. The inability to express MarR homologs in B. fragilis influenced antimicrobial susceptibility and rendered both mutant strains more susceptible to teicoplanin, tetracyclin, imipenem, meropenem, cefotaxin, cefepime, levofloxacin and metronidazol (Table 1). The MIC for polymyxin remained unchanged and strain LE64 was more susceptible to linezolid than the wild type strain.
DiscussionIt is estimated that only 1% of all Bacteroides spp. present in the gastrointestinal tract belong to the B. fragilis species. Despite being a minor component of the microbiota, it has important roles in the gut, particularly in the development of the immune system.26 In cases of disbiosis or rupture of the integrity of the intestinal mucosa it also emerges as an opportunistic pathogen and it is the most commonly isolated strict anaerobic bacteria from human infection.2 This ambiguous interaction with the human host may be explained in part by the expression of several virulence factors, including the CPC, adhesins and secreted hydrolytic enzymes.4 Although the complete details of the transition from a symbiotic to a pathogenic state are not fully understood, the capacity to survive the hostile oxygenated environment of the peritoneal cavity and resist to antimicrobial drugs certainly play a decisive role.
It has been demonstrated that upon exposure to oxygen, B. fragilis mounts a stress response that may affect up to 45% of its genome.7 Interestingly, the oxidative stress response also affects the expression of virulence factors27 indicating that it may work as an environmental sensor for the transition from the anaerobic gut lumen to the oxidized peritoneal cavity. Recently we demonstrated that the insertion of a DNA fragment in the gene locus BF638R_0571 disrupted the expression of a MarR homolog in B. fragilis leading to an altered response to oxidative stress.10 This gene product was renamed BmoR, and mutant strains unable to express this protein were more susceptible to oxidative stress than their parental strain. The 638R strain used in the aforementioned study carries at least two other homologs of MarR proteins whose function are not known. We constructed two single mutant strains, each carrying a disruption in one of the two remaining MarR homologs and analyzed the subsequent phenotype. Susceptibility to oxygen was increased after long-term exposure (72h) in the mutant strains, but not to hydrogen peroxide. Similarly, BmoR was shown to respond to oxygen stress, with only a modest response to hydrogen peroxide.10 Since hydrogen peroxide is one of the major reactive species generated by oxygen exposure, the lack of response to this compound could mean that MarR homologs in the species are not associated with the detoxification of oxygen, but rather with a metabolic pathway related to redox balance in the cell. Anaerobic bacteria are known to contain separate response mechanisms for different sources of oxidative stress.8,28,29 The regulator OxyR in B. fragilis for example regulates a complex global response that leads to detoxification of hydrogen peroxide. Although OxyR mutants are more susceptible to atmospheric oxygen than wild type strains, the stress response orchestrated by OxyR is directed at preventing a collateral damage caused by oxidative stress, the accumulation of H2O2 in the cell.5,8 Unfortunately, besides OxyR there are few studies associated with regulation of oxidative stress response and several redox proteins that have no known regulator associated with, like the thioredoxins that contribute for the oxidative stress response by providing reducing power, which could be controlled by, for example, MarR homologs.
MarR family proteins are implicated in resistance to biliary salts in Gram negative pathogens such as Salmonella enteritidis.30 Direct binding of the bile salt deoxycholate to MarR was demonstrated to inhibit DNA binding and to de-repress the marRAB operon, but surprisingly, this effect does not increase resistance to bile salts in S. enteritidis. Similarly, we did not observe any change in susceptibility to bile salts in the B. fragilis mutant strains. Since B. fragilis is well adapted to the intestinal environment, it is not surprising that redundant mechanisms of regulation for bile salts susceptibility are in place. In Gram negatives, regulation of virulence genes is also affected by MarR homologs.15 This global regulation profile reflects the need to couple environmental sensing and virulence gene expression in bacterial pathogens. MarR homologs regulate the expression of surface structures such as polysaccharide capsule in Bordetella bronchiseptica31 and fimbriae in E. coli with a consequent impairment in formation of biofilm.32 Again, in B. fragilis we did not observe any effect on biofilm formation. In vitro biofilm assays are limited by the inconvenience of replicating the conditions found in the gut epithelium. Even with the mutation in marR homologs, artificial media and physical support (polystyrene microplates) may be permissive for biofilm accumulation.
Despite modest results found in the previous phenotypical tests, the effect of the abolishment of the expression of MarR homologs in antimicrobial susceptibility was noticeable. MarR was first described as the repressor of an operon related to multi-drug resistance in E. coli, and mutations in MarR in E. coli leads to an increased resistance profile.33,34 The increase in resistance in this species is associated with the expression of efflux systems negatively regulated by MarR.35 Conversely, in our experiments, disruption of the expression of MarR homologs leads to decreased resistance to several antimicrobial drugs, including drugs from different classes and with different targets in the cell. This result points to a defect on a general mechanism of resistance, such as efflux pumps or membrane permeability. Several explanations may account for this contradictory effect. First, despite the homology between marR in B. fragilis and E. coli, the genetic contexts are different in these species. There is no homolog to MarA or MarB downstream MarR homologs in B. fragilis for example. Thus, the genes controlled by this regulator may be in a different location in the chromosome. Although MarR was shown to regulate the expression of efflux systems in some bacterial species,35 in others it controls the expression of virulence genes and outer membrane proteins15 as well as proteins that modify the architecture of the cell envelope.31 It is possible that outer membrane porins involved in the entrance of antibiotics in the cell are upregulated in our mutants, allowing more drugs to reach their target. And finally, there is a possible epigenetic effect caused by the insertion of a plasmid in the bacterial chromosome leading to downregulation of genes downstream of our target genes, an opposite effect to what would be observed in the event of a lack of a repressor. This later effect is currently being investigated in our lab, but the lack of appropriate molecular tools suitable for strict anaerobic species such as B. fragilis is frustrating.
Although many answers still remain elusive, our work is the first to study transcriptional regulators of the MarR family in the strict anaerobe B. fragilis in relation to antimicrobial resistance. There is a clear effect observed in antibiotic resistance associated with disruption of the expression of MarR homologs. This indicates a participation of these proteins in the regulation of resistance factors. The change in the resistance profile was not specific for a particular class of antibiotics, and as mentioned, a general mechanism such as membrane permeability may be involved. We did not observe significant alterations in other aspects of the physiology of the bacterium such as biliary salts and biofilm formation. We hypothesized that, as observed with other bacterial species, genes involved in membrane permeability are modulated by MarR homologs in B. fragilis. Understanding these mechanisms is paramount for the development of new strategies to fight multi-drug resistant infections caused by this opportunistic pathogen.
Conflicts of interestThe authors declare no conflicts of interest.
The authors want to thank Mr. Joaquim da Silva for technical support. This work was funded by Fundação de Amparo a Pesquisa do Rio de Janeiro (FAPERJ), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES).