A total of 276 endophytic bacteria were isolated from the root nodules of soybean (Glycine max L.) grown in 14 sites in Henan Province, China. The inhibitory activity of these bacteria against pathogenic fungus Phytophthora sojae 01 was screened in vitro. Six strains with more than 63% inhibitory activities were further characterized through optical epifluorescence microscopic observation, sequencing, and phylogenetic analysis of 16S rRNA gene, potential plant growth-promoting properties analysis, and plant inoculation assay. On the basis of the phylogeny of 16S rRNA genes, the six endophytic antagonists were identified as belonging to five genera: Enterobacter, Acinetobacter, Pseudomonas, Ochrobactrum, and Bacillus. The strain Acinetobacter calcoaceticus DD161 had the strongest inhibitory activity (71.14%) against the P. sojae 01, which caused morphological abnormal changes of fungal mycelia; such changes include fracture, lysis, formation of a protoplast ball at the end of hyphae, and split ends. Except for Ochrobactrum haematophilum DD234, other antagonistic strains showed the capacity to produce siderophore, indole acetic acid, and nitrogen fixation activity. Regression analysis suggested a significant positive correlation between siderophore production and inhibition ratio against P. sojae 01. This study demonstrated that nodule endophytic bacteria are important resources for searching for inhibitors specific to the fungi and for promoting effects for soybean seedlings.
The root nodules of legume plants are symbiotic organs induced by soil bacteria known as rhizobia. As part of the root system, root nodules harbor symbiotic bacteria and many endophytes, including Agrobacterium tumefacien,1A. rhizogenes,2Phyllobacterium, Stenotrophomonas, Enterobacteriaceae,3Bacillus species,4Bacillus, Bordetella, Curtobacterium, and Pantoea.5 Aside from their diversity, which has been studied extensively, the effect of nodule endophytes on host legumes was revealed. The nodule endophytic Agrobacterium strains specifically inhibit the nodulation of Rhizobium gallicum on the common bean (Phaseolus vulgaris L.)6 or facilitate the unspecific nodulation of Sinorhizobium meliloti on woody legumes.7 Some nodule endophytes that belong to Bacillus, Bordetella, Curtobacterium, or A. rhizogenes could promote the growth or nodulation of red clover.5 These phenomena are similar to that of endophytes isolated from other parts of plants and could benefit host plants by producing phytohormones, 1-aminocyclopropane-1-carboxylase (ACC) deaminase, and antibiotic compounds, as well as by fixing nitrogen, solubilizing phosphate, or suppressing phytopathogens through the competence of invasion sites.8–11 Owing to the above mentioned advantages, endophytes are considered novel resources in the biocontrol of plant diseases and in the promotion of plant growth.12–14
As a major legume crop, soybean (Glycine max L.) plays an important role in sustainable agriculture and in the economy of many countries. Soybean has a great nitrogen-fixing ability due to its symbiosis with rhizobia in root nodules. The presence of Bradyrhizobium japonicum, B. liaoningense, B. yuanmingense, B. elkanii,15,16B. huanghuaihaiense,17B. daqingense,18B. pachyrhizi, B. iriomotense, B. canariense,19Sinorhizobium fredii, and S. sojae20 has been reported in China, which is the center of origin of soybean.21,22 Similar to other plants, endophytic bacteria have been isolated from different parts of soybean,19,23–26 and some of these parts showed antagonistic and growth-promoting potential.27–29 Diverse endophytic bacteria, including Pantoea, Serratia, Acinetobacter, Bacillus, Agrobacterium, and Burkholderia, have also been isolated from soybean nodules.30 However, antagonistic endophytic bacteria within nodules of soybean for P. sojae in Henan Province have not been sufficiently studied.
On the basis of the above mentioned background knowledge and considering the nodule endophytes as a new bacteria resource with potential in biotechnology, we conducted this study (1) to screen antagonistic endophytic bacteria from soybean nodules against P. sojae; (2) to explore the potential plant-beneficial traits of endophytic bacteria; and (3) to assay the seedling growth response of soybean to the inoculation of endophytic bacteria.
Materials and methodsCollection of root nodules, soil samples, phytopathogenic fungus, and soybean seedsNodules from cultivated soybean were collected from July to August 2012, when the plants were blooming. Samples were obtained from fields of 14 sites subordinate to 9 districts of Henan Province, China (map available as Supplementary Fig. 1).21,22 Three healthy root nodules with similar sizes were excised from the lateral roots of each plant. Soil debris was brushed away from the nodules, and the nodules were stored in sterile plastic bags at 4°C until they were processed for isolation within 24h.
In each site, soil cores were sampled at five locations with a depth of 15–20cm and 5cm away from the taproots, which were bulked and thoroughly mixed to form composite samples. Soil samples were stored in loosely tied plastic bags at 4°C. Soil texture was defined according to the international institution triangle coordinate graph, and soil pH was determined as described in Zhao et al.31
A phytopathogenic fungus, P. sojae 01, was provided by the College of Life Sciences of Northwest A & F University in China and was incubated on potato dextrose agar plate (PDA: extract of 200g potato, 20g of glucose, 18g of agar, 1L of distilled water) at 30°C for 3 days and maintained at 4°C for temporary storage.
The seeds of soybean (G. max L.) cultivar Zhonghuang 13, which is the principle cultivar used in the sampling region, were bred by the Institute of Crop Sciences of the Chinese Academy of Agricultural Sciences.
Isolation and purification of soybean nodule endophytesBacteria were isolated from root nodules according to a standard method as described by Ma et al.32 and Miller et al.33 A single colony of the isolate was repeatedly streaked on the same medium and examined with a microscope. Pure cultures were preserved on plates at 4°C for temporary storage or in sterile vials with 30% (v/v) glycerol for long-term storage at −80°C. To confirm if the surface sterilization process was successful, several surface-sterilized nodules were rolled over nutrient agar (NA) plates and aliquots of water from final rinse solutions and then plated onto NA plates.34 Plates without any contaminants were considered effectively surface-sterilized, and the corresponding plates were used for the isolation of endophytes.
Screening of antagonistic endophytic bacteriaThe antifungal activity of endophytes against pathogenic fungus P. sojae 01 was detected by using the point inoculation method.35 Spores of fungal cultures were inoculated on PDA plates, and a small block of agar with fungal mycelia cut with a sterile puncher (Ø=4mm) was placed in the center of a fresh plate. Tested strains were spot inoculated on the edge of PDA plates approximately 25mm from the center. After incubation at 28°C for 7 days, the inhibition zone was measured. Fungal mycelia that were cultivated without inoculation were included as control.36 Experiments were performed in triplicate for each bacterial isolate.
Secondary screening of antifungal activity was performed similar to the primary screening method, but bacteria were spot inoculated as bacterial suspension (OD600≈1). Antagonistic activities were evaluated by measuring inhibition zones between pathogens and tested bacteria.
Microscopic observation of phytopathogenic fungi myceliaTo determine the effect of endophytic bacteria on pathogenic fungus, treated and untreated pathogenic fungi were cultured for 2 days on PDA medium. The morphological changes of pathogenic fungus caused by endophytes were examined under an optical epifluorescence microscope (BX50 Olympus) at 200-fold magnification and compared with the structures of the control groups. The mycelium of each pathogenic fungus on the growth PDA medium was directly examined and photographed from the plates by using a digital camera (Olympus).
Sequencing and phylogenetic analysisThe total genomic DNA was extracted from the culture of nodule isolates by using the previous method.37 The 16S rRNA gene was amplified from the genomic DNA by PCR with the universal forward primer P1 (5′-CGGGAT CCA GAG TTT GAT CCT GGC TCA GAA CGA ACG CT-3′) and reverse primer P6 (5′-CGGGAT CCT ACGGCT ACC TTG TTA CGA CTT CAC CCC-3′), respectively, which corresponded to the positions of 8–37bp and 1479–1506bp in Escherichia coli 16S rRNA gene.38 An aliquot of PCR product of isolates was directly sequenced by Sangon Biotech (Shanghai) Co., Ltd. using the same primers mentioned above. Acquired and related sequences were matched with ClustalX1.81 software, imported into Bioedit 4.8.4, and manually corrected. A phylogenetic tree was constructed using the Jukes–Cantor model and the neighbor-joining method39 in TREECON package (version 1.3b).40 The similarity of each tested strain was computed by using the DNAMAN application (version 6.0.3.40, Lynnon Corporation). The acquired 16S rRNA gene sequences were submitted to NCBI GenBank (http://www.ncbi.nlm.nih.gov/). The GenBank accession numbers of the sequences obtained in this study are KF843714–KF843719.
Siderophore productionBacteria were cultured in Luria–Bertani (LB) broth at 30°C with shaking at 130rpm until the exponential growth phase (OD600≈1) was achieved. The production of siderophores by the bacteria was determined according to the chrome azurol-S (CAS) analytical method.41 The supernatant was obtained by centrifugation at 9000×g for 10min and then mixed with 1mL of CAS assay solution.42 A medium mixed with the CAS assay solution at a 1:1 ratio was included as blank control, and the difference of OD630 between the treatment and blank was estimated as values of siderophore production.43 Experiments were performed in triplicate.
Nitrogen fixation and nifH gene amplificationThe fixation of atmospheric nitrogen by the bacterium was tested qualitatively using Ashby's N-free medium (NFM: 10g of mannitol, 0.2g of KH2PO4, 0.2g of MgSO4·7H2O, 0.2g of NaCl, 0.1g of CaSO4·2H2O, 5g of CaCO3, pH 7.0–7.5, 1.8g agar in lL of distilled water).23 Plates were incubated at 28°C for 3 days, and strains that grew normally in NFM media were defined as candidates of nitrogen fixers.30,44 Experiments were performed in triplicate. The forward primer nifH40F (5′-GGN ATC GGC AAG TCS ACS AC-3′), reverse primernifH817R (5′-TCR AMC AGC ATG TCC TCS AGC TC-3′),and the procedure described by Vinuesa et al.45 were used for nifH gene specific amplification by PCR. PCR products were separated by horizontal electrophoresis in 1% (w/v) agarose gels, and patterns were visualized.
Indole acetic acid (IAA) productionIAA production was estimated by inoculating a bacterial suspension (1×108cfumL−1) in 10mL LB broth that contained l-tryptophan (100μgmL−1) and shaken at 30°C for 72h in the dark. Five milliliters of each culture were centrifuged (20min, 6000×g), and IAA production was measured as indolic compounds in 2mL of supernatant mixed with 2mL of Salkowski reagent, and the absorbance was read at 535nm after 30min incubation in the dark.46 A standard curve was used for calibration to quantify. Three replicates were performed for each IAA synthesis measurement.
Plant inoculation studies with endophytic bacteriaAntifungal endophytic bacteria were cultured in TY agar medium at 30°C to the mid-log phase.47 Cells were pelleted by centrifugation (3440×g, 10min at 4°C), washed twice with a sterile saline solution, and prepared for bacterial suspensions (approximately 108cfumL−1). The treatment of soybean seeds was the same as the surface sterilization of nodules.31 In each sterile Petri dish, 30 surface-sterilized seeds were placed separately on moist filter paper for germination at 28°C. Germinated seeds were immersed in bacterial suspension for 8h. Experiments were conducted in triplicate. The control was immersed with sterile water.
Inoculated seedlings were sown in pots filled with 190g sterilized vermiculite and then incubated in the greenhouse with a photoperiod of 16h daylight at 22°C, a night temperature of 20°C, and 65% relative humidity. After the first main leaf appeared, each seedling was inoculated with 108cfu of the tested strain every week, and sterilized water was poured every 3 days to maintain relative humidity.47 Seedlings without inoculation were included as blank control. Plants were harvested after 6 weeks, and root length, fresh weight, and chlorophyll content were determined.
Statistical analysisData collected from growth promotion and endophytic inoculation experiments were examined with ANOVA using the IBM SPSS 17.0 package (by the Data Theory Scaling System Group, Faculty of Social and Behavioral Sciences, Leiden University, The Netherlands). The effects of six endophytic bacteria on shoot length, root length, fresh weight, and chlorophyll content of soybean seedlings were analyzed with GraphPad Prism 5.01 software.
ResultsIsolation and screening of antagonistic bacteriaA total of 276 bacterial isolates were obtained, of which 31 showed significant inhibition (inhibition ratio >42%) against P. sojae 01 in the initial and secondary screenings (Supplementary Table S1). Six isolates that showed more than 63% inhibition for mycelial growth of P. sojae 01 on PDA plate were selected for further study. These isolates were DD161 (71.14% inhibition), DD176 (70.30%), DD198 (68.43%), DD222 (64.32%), DD201 (63.40%), and DD234 (63.16%).
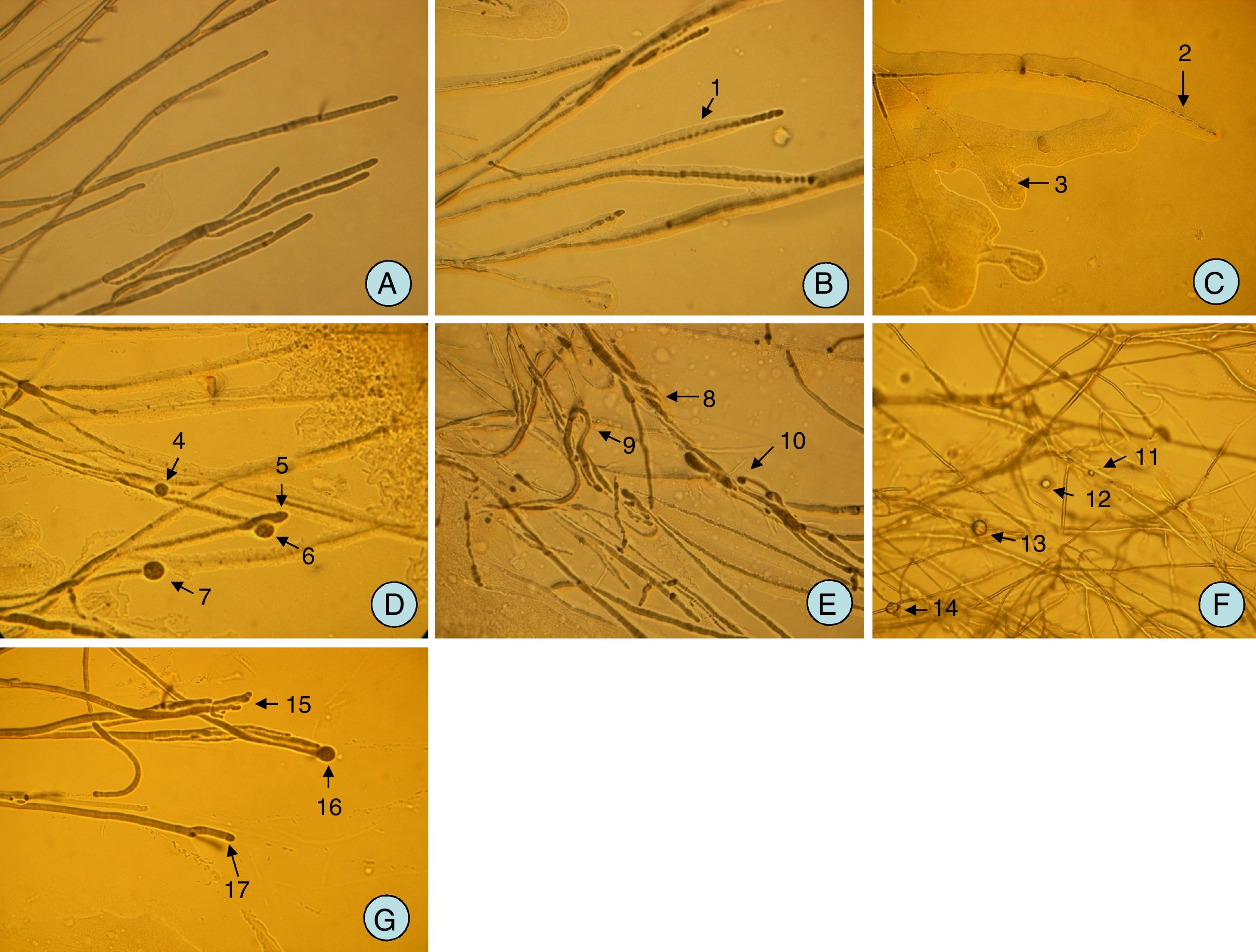
Microscopic observation of phytopathogenic fungusThe colonies of pathogenic fungus were more inhibited after 4 days of culturing with endophytic bacteria on PDA medium compared with the control. Microscopic observation showed that the fungal mycelia presented morphological changes in the treatment of endophytic bacteria. The treated fungus became fractured (Fig. 1B) orlysed (Fig. 1C) and were wrapped with biofilm formed by the bacteria (Fig. 1B and C) unlike the control (Fig. 1A). In addition, for the mycelia treated by endophytic bacteria, the hyphae ends became protoplast balls (Fig. 1D and G) or split (Fig. 1G) even though they were not wrapped by biofilm. Some aerial hyphae showed sarciniform wrapped around each other (Fig. 1E) and twisted (Fig. 1E), as well as a fractured and spherical protoplastend. Furthermore, some aerial hyphae became thin, transparent, and bent, and formed transparent liquid droplets (Fig. 1F) under the action of endophytic bacteria.
Morphological changes of the mycelia of plant pathogenic fungi upon interaction with endophytes isolated from soybean nodules. (A) Normal mycelia of Phytophthora sojae 01(CK); (B) mycelia became wrapped with biofilm formed by endophytic bacteria DD222 (1); (C) mycelia became fractured (2), lysis (3) under effect by endophytic bacteria DD161; (D) hyphae end became protoplast concentration and formed a ball (4, 5, 6, and 7) for mycelia unwrapped by biofilm under the action of DD201; (E) some aerial hyphae showed sarciniform (8) wrapped around each other (8) and twisted (9, 10) under the action of DD198. (F) Aerial hyphae became thin, transparent, and bent, and formed transparent liquid droplets (11, 12, 13, and 14) under the action of endophytic bacteria DD234; (G) hyphae end became split ends (15, 17) and protoplast concentration appeared spherical (16) under the action of endophytic bacteria DD176.
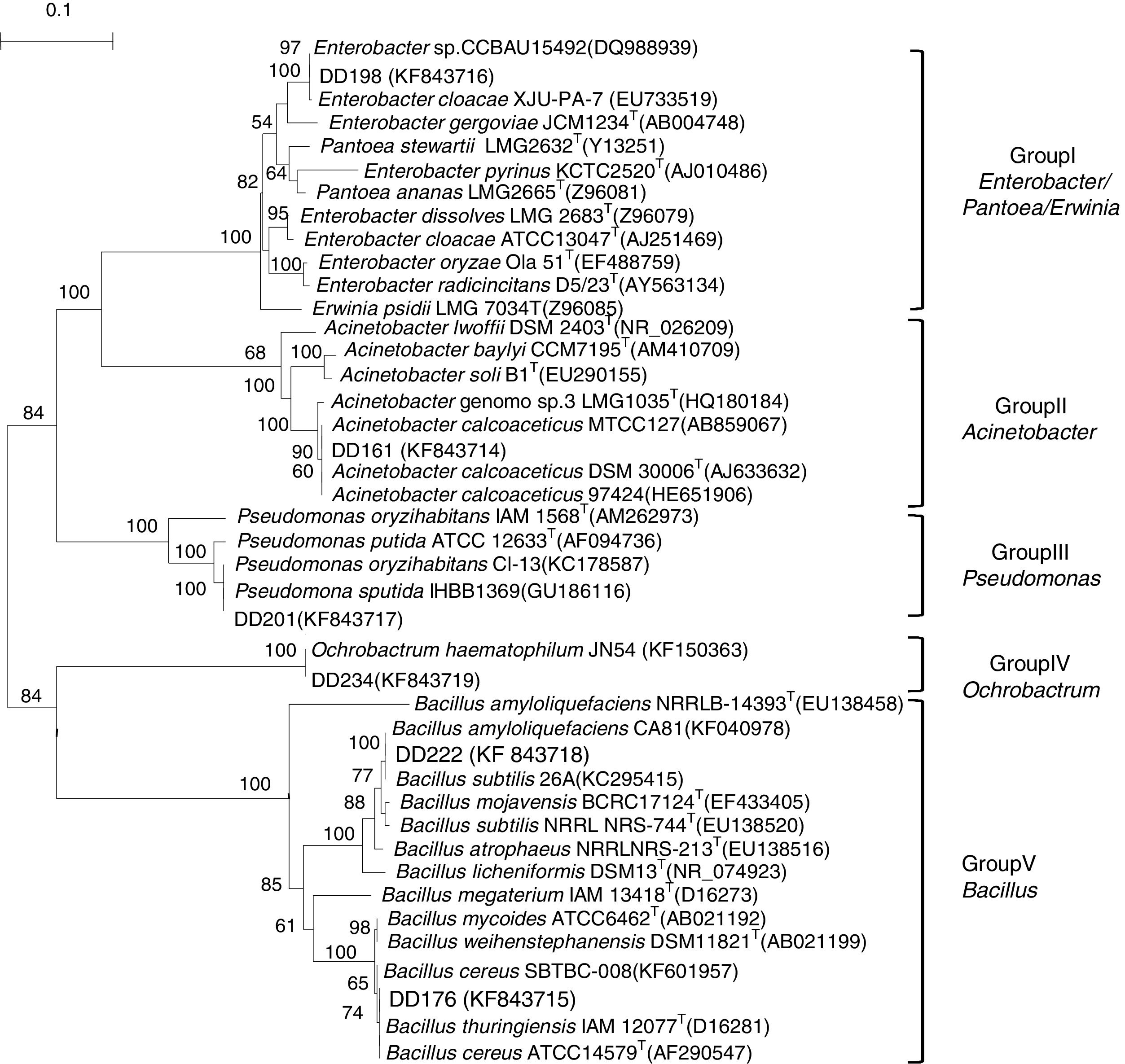
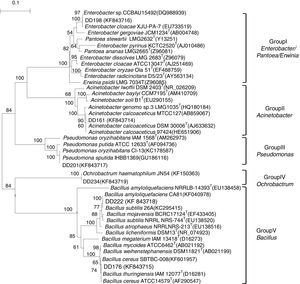
The phylogeny of 16S rRNA genes indicated that six endophytic antagonists belonged to five genera, as shown in Fig. 2 and Table 1. DD198 showed 99.9% similarity with Enterobacter cloacae XJU-PA-7 (EU733519). DD161 had 100% sequence similarities with Acinetobacter calcoaceticus. DD201 was 100% similar to Pseudomonas putida, DD234 was 100% similar to Ochrobactrum haematophilum, and DD222 and DD176 presented 100% similarities with Bacillus amyloliquefaciens and Bacillus cereus, respectively.
Neighbor joining tree based on alignment of nucleotide sequences of the 16S rRNA gene from tested strains (shown in bold) and reference strains. GenBank accession numbers were placed in parentheses. Bootstrap values greater than 50% were indicated. Scale bar represents the number of substitutions per site.
Phylogenetic similarity and plant-growth promoting properties of endophytic bacteria.
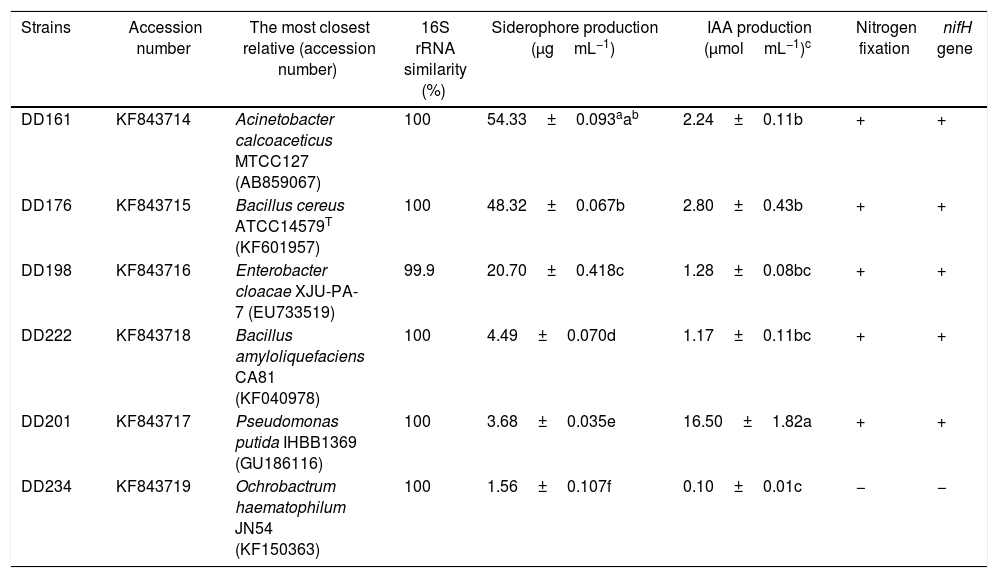
| Strains | Accession number | The most closest relative (accession number) | 16S rRNA similarity (%) | Siderophore production (μgmL−1) | IAA production (μmolmL−1)c | Nitrogen fixation | nifH gene |
|---|---|---|---|---|---|---|---|
| DD161 | KF843714 | Acinetobacter calcoaceticus MTCC127 (AB859067) | 100 | 54.33±0.093aab | 2.24±0.11b | + | + |
| DD176 | KF843715 | Bacillus cereus ATCC14579T (KF601957) | 100 | 48.32±0.067b | 2.80±0.43b | + | + |
| DD198 | KF843716 | Enterobacter cloacae XJU-PA-7 (EU733519) | 99.9 | 20.70±0.418c | 1.28±0.08bc | + | + |
| DD222 | KF843718 | Bacillus amyloliquefaciens CA81 (KF040978) | 100 | 4.49±0.070d | 1.17±0.11bc | + | + |
| DD201 | KF843717 | Pseudomonas putida IHBB1369 (GU186116) | 100 | 3.68±0.035e | 16.50±1.82a | + | + |
| DD234 | KF843719 | Ochrobactrum haematophilum JN54 (KF150363) | 100 | 1.56±0.107f | 0.10±0.01c | − | − |
Table 1 summarizes the results of PGP trait evaluation in vitro. Except for DD234, the other five strains showed the capacity to produce siderophore and IAA, as well as the capacity to fix nitrogen. Different biosyntheses of siderophores were found among the strains. DD161, DD176, and DD198 produced 54.3, 48.3, and 20.7μgmL−1 of siderophores, respectively, while DD222, DD201, and DD234 produced less than 5μgmL−1 of siderophores in the same experimental conditions. Regression analysis showed a significant positive correlation between siderophore production and inhibition ratioagainst P. sojae 01 (R=0.9643, p=0.0019<0.05) (details available in Supplementary Fig. S2). IAA production of DD201 was significantly higher (16.5μgmL−1) than that of the other five (<3μgmL−1).
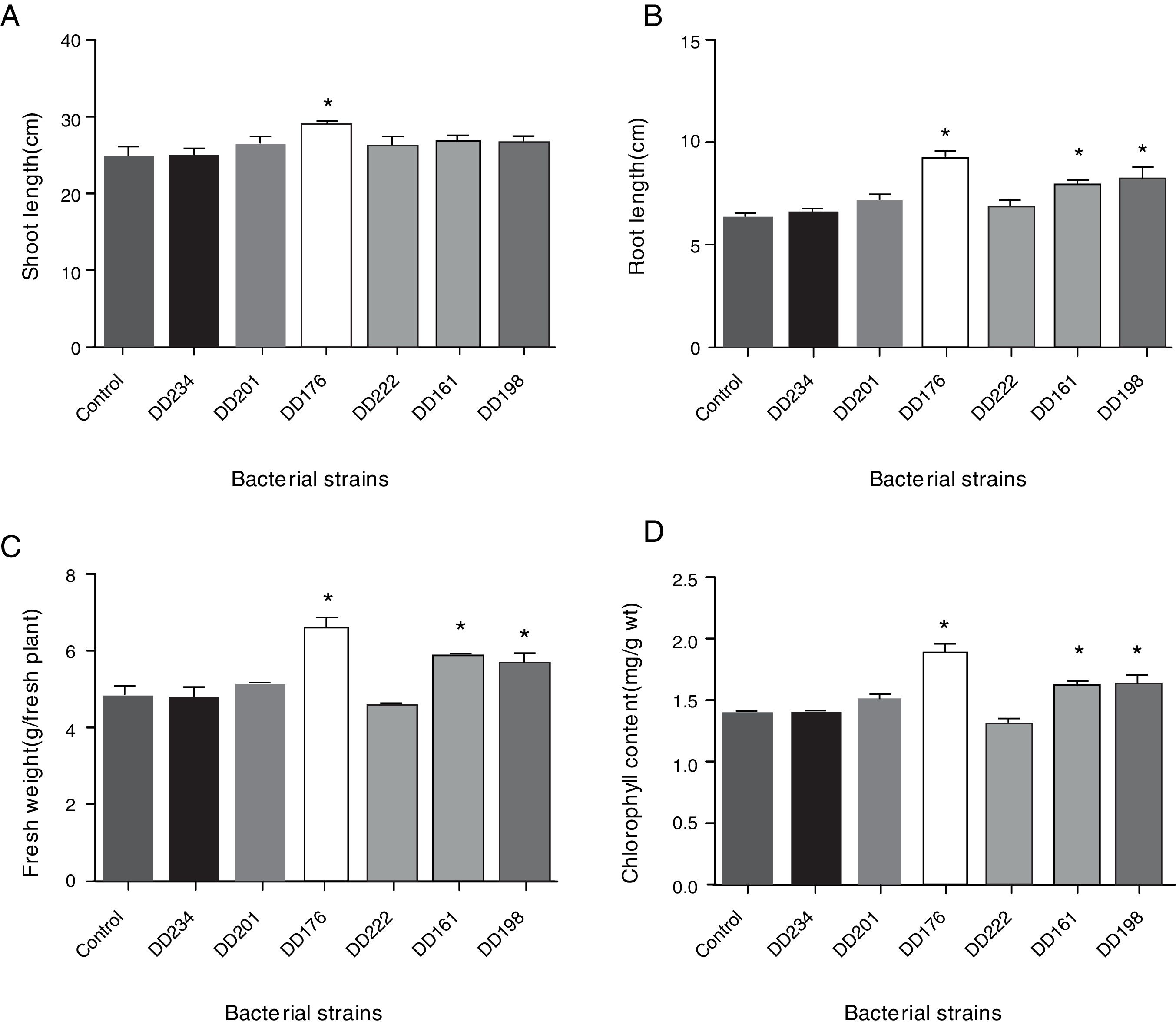
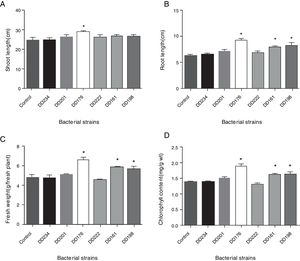
Seedling growth response to the inoculation of endophytic bacteriaThe results in Fig. 3 showed that inoculations with DD176 significantly increased (19.2%, p<0.05) the shoot length of soybean seedlings. Inoculations with DD176, DD161, and DD198 resulted in a significant increase in root length (38.32%, 36.23%, and 29.82%, respectively) (p<0.05), fresh weight of plants (36.45%, 20.47%, and 17.00%, respectively) (p<0.05), and chlorophyll content (36.73%, 17.09%, and 13.75%, respectively). Overall, these results showed that inoculation of the tested endophytic bacteria significantly improved the growth of soybean seedlings.
Effect of six endophytic bacteria on shoot length (A), root length (B), fresh weight (C), and chlorophyll content (D) of soybean seedlings. Each value is the mean of 10 replicates. Bars represent the standard deviations of mean. Statistical significance was determined at p<0.05 according to Tukey's test. Asterisk represents significant difference.
Currently, endophytic microorganisms are believed to be an important bioresource for modern agriculture48 because of the beneficial effects of endophytes on plant growth promotion, biocontrol, and disease resistance. As a part of the root system, root nodules are also a habitat for endophytes. However, this habitat is different from other parts of plants because the endophytes in this habitat have to compete and co-exist with symbiotic bacteria and help the plants through certain mechanisms. In this study, a significant inhibitory activity against pathogenic fungus P. sojae 01 was found among 11.2% (31/276) of the nodule endophytic bacteria (Table 1 and Supplementary Table S1). The high proportion of fungal antagonistic bacteria in the root nodules revealed in this study and in previous studies49 demonstrated that antagonistic activity might be a universal mechanism through which nodule endophytic bacteria can help host plants.
In addition to the antagonism against pathogenic fungi, all six strains produced siderophores and IAA, while five strains were capable of fixing nitrogen. These results demonstrated that nodule endophytic bacteria have diverse functions in the inhibition of phytopathogens and in promoting growth.
Currently, several possible mechanisms are suggested for the inhibition of phytopathogens by endophytic bacteria: (1) competition with pathogens for the ecological niche/substrate (siderophores) in the rhizosphere; (2) production of antibiotics (cyclic lipopeptides, iturin, fengycin)50 and antifungal substances (2,4-diacetylphloroglucinol); (3) production of extracellular chitinase and laminarinase to lyse fungal cells51 and degrade fusaric acid produced by fungal pathogens52; and (4) production of volatile organic compounds(such as 2,3-butanediol and acetoin, which act as signaling molecules to mediate plant–microbe interactions),53 which could strongly inhibit pathogen growth on different hosts54 and elicit plant growth by induced systemic resistance (ISR).50,55 The results obtained in the current study might indicate evidence of the first and the third mechanisms.
The high correlation (R2=0.93) of siderophores and the fungal inhibition of nodule endophytic bacteria in this study (Supplementary Fig. S2) supported the idea that the strong ferrous absorption by endophytic bacteria might be a mechanism for the inhibition of fungal growth. Meanwhile, the formation of biofilm on the hyphae and the morphological changes of the mycelia of the target fungus (Fig. 1) showed that antifungal substances and fungal cell-lysing enzymes might be produced by the endophytic bacteria, as reported by Mauch et al.51 These results demonstrated that nodule endophytic bacteria are important resources for searching for inhibitors specific to the fungi but without negative effects on symbiotic bacteria.
In accordance with the phylogeny of 16S rRNA genes (Fig. 2), the six most efficient (>63% inhibition) antagonistic strains, which were preliminarily identified as belonging to five genera (Table 1), demonstrated again that the root nodules could be occupied by diverse bacteria. These findings supported the results of other studies.3,30,56,57 All these five genera, namely, Acinetobacter, Bacillus, Enterobacter, Ochrobactrum, and Pseudomonas, have been reported previously as nodule endophytes of different legumes, including soybean.3,58–60 However, these endophytes were isolated from soybean nodules collected from different regions of Henan Province, thereby suggesting the antagonistic effect of soybean endophytes on pathogenic fungus of diverse geographical sources and species. This diversity may be attributed to multiple symbiotic relationships in the particular region (the original area of soybean).21,22 During a long evolution period, soybean, rhizobia, and endophytes formed a multiple symbiotic relationship, and soybean provides nutrients and a suitable environment for symbiotic and endophytic bacteria. Rhizobia provided nitrogen nutrition for plants and endophytes, while endophytes strengthened the resistance of plants and symbiotic bacteria against pathogens and bad environmental factors.
In this study, two of the antifungal endophytic bacteria (DD176 and DD222) were identified as Bacillus sp. Bacillus is one of the most abundant rhizosphere bacteria and nodule endophytes.34,58,60 These bacteria could improve the yields of various crops61–63 by stimulating plant growth (with hormones) and improving nutrient supply (with phosphate-solubilizing siderophores)or by antagonism against phytopathogens through protease or cellulose production.64–66 Our results showed that B. sp.DD176 and B. sp.DD222 produced siderophore and hormones (IAA) in addition to effectively inhibiting the pathogenic fungus (Table 1). The formation of biofilm and the accompanying morphological changes in the mycelia (Fig. 1) supported the idea that Bacillus spp. could produce several types of enzymes to degrade fungal cell walls, which resulted in a protoplast ball or split ends of the mycelia. Furthermore, B. amyloliquefaciens strains are characterized by high rhizosphere competence and a significant genetic apparatus devoted to the biosynthesis of a wide range of substances with antibiotic activity.55,67–71 All previous studies and our study demonstrated that nodule endophytic Bacillus strains are valuable candidates for exploring biofertilizers.
The isolate DD198 was identified as Enterobacter cloacae, which is a rhizophere bacterium34,58,60,72 that produces phytohormones, such as ethylene, auxins, cytokinins,65 siderophores,63,73 and fixes nitrogen.72 In the present study, strain E. cloacae DD198 showed significant inhibitory activity against P. sojae 01 in vitro and promoted effects for wheat seedlings with inoculation treatment.
In our study, A. calcoaceticus DD161 possesses the strongest ability to produce siderophores and inhibit the growth of pathogenic fungus, as well as synthesize IAA. The comprehensive effect indicated that the growth of soybean seedling inoculated with DD161 was significantly improved (Fig. 3). Interestingly, only a few reports showed that A. calcoaceticus strains indicated both nitrogen fixation activity and inhibition effect aside from their PGPR activity. Previous reports showed that A. calcoaceticus isolated from rhizosphere of wheat74,75 could synthesize IAA from tryptophan and produce siderophores and phosphate-solubilizing organic acids. Therefore, the A. calcoaceticus strain may improve crop growth and yield on the basis of its biocontrol activity, siderophore production, and nitrogen fixation.76
Isolate DD201 was identified as Pseudomonas sp., which is an opportunistic bacterium found in terrestrial and aquatic environments, and indicated biotechnological behaviors.77 In this context, P. sp. DD201 showed the highest IAA production (16.5μmolmL−1) but did not significantly improve the growth of soybean seedlings in inoculation tests (Fig. 3). Previous studies confirmed that IAA could promote plant grow that low concentration and inhibit root growth at high concentration.78–80 Presumably, PGP bacteria use IAA as a part of their colonization strategy and as a signal molecule in bacteria–host communication.66 These functions might explain the reason for the production of IAA that was common among our six endophytic strains (Table 1).
This study proved that fungal antagonistic strain DD234 was O. haematophilum (Table 1). Ochrobactrum isolates could assist plant nutrient uptake from the soil and prevent plant diseases.81 The plant growth-promoting characteristics of siderophore production, IAA production, and phosphate solubilization were found in some strains of this genus82 and might be the mechanism for increasing host plant growth.83
Conflicts of interestWe declare that we have no conflict of interest to this work.
This work was supported by projects from the National Science Foundation of China (U1204301), the Foundation for University Key Teacher by the Ministry of Education of Henan Province (2012GGJS166) and the University Key Scientific Research Project of Henan Province (17A180011).