This study examined the antimicrobial susceptibility patterns and clonal complex (CC) characteristics of serogroup 6 Streptococcus pneumoniae isolates collected from children in Beijing, China, between 1997 and 2016. Serotypes were determined using the Quellung reaction, and the antimicrobial susceptibility profiles of the isolates were determined using the disc-diffusion method or by E-test. Sequence types (STs) were assigned based on multilocus sequence typing. A total of 250 isolates were examined, with 55.2%, 30.0%, 12.8%, and 2.0% of isolates identified as serotypes 6A, 6B, 6C, and 6D, respectively. All of the isolates were susceptible to levofloxacin and vancomycin, and the non-suceptibitility rate to penicillin was 41.6%. Eighty-two distinct STs, assigned to 13 CCs and 28 singletons, were identified. CC982 was the most prevalent CC amongst serotype 6A isolates (34%), followed by CC9789 and CC3173. Amongst serotype 6B isolates, CC90 and CC4542 were the most common, accounting for 25.3% and 14.7% of isolates respectively. Over the study period, the prevalence of CC982, CC4542, and CC4536 isolates showing susceptibility to penicillin and cefuroxime decreased, and the proportion of CC3173, CC9789, CC855, and CC902 isolates showing non-susceptibility to these two antibiotics increased.
Streptococcus pneumoniae, the most common cause of bacterial pneumonia, can cause a series of invasive and non-invasive diseases, and seriously threatens the health of children worldwide. Using the Danish typing system based on differences in the polysaccharide capsule, more than 90 S. pneumoniae serotypes have been detected. Serogroup 6, comprising serotypes 6A, 6B, 6C, and 6D, is one of the most prevalent serogroups worldwide.1–3
The first pneumococcal conjugate vaccine (PCV), PCV7, offered protection against seven serotypes (4, 6B, 9V, 14, 18C, 19F, and 23F) and was licensed in 2000. By 2015, PCV7 had been replaced with the higher valent vaccines PCV10 (PCV7+1, 5, and 7F) and PCV13 (PCV10+19A, 6A, and 3), which were introduced in over 130 countries.4,5 Research into the efficacy of PCVs showed that their widespread availability decreased nasopharyngeal carriage rates and infections caused by vaccine serotypes.6–8 However, a serotype replacement phenomenon appeared with the introduction of PCVs, whereby the prevalence of non-vaccine serotypes increased with concomitant decreases in vaccine serotypes.9–12 While there is no universal immunization program for PCVs in China, PCV7 was licensed in 2008 by the private sector, and was withdrawn at the end of 2014. Despite this, an investigation performed in 2011 showed that the vaccination coverage of PCV7 in children under 2 years old was only 9.91%.13 PCV13 was not introduced in China until April 2017, and serogroup 6 pneumococcal strains remain prevalent.14–16
The aim of the current study was to examine the antimicrobial susceptibility profiles and clonal complex characteristics of serogroup 6 S. pneumoniae isolates collected from children in Beijing between 1997 and 2016, and to explore the relationship between antibiotic resistance and clonal complex.
Materials and methodsBacterial strainsA total of 1865 S. pneumoniae isolates were cultured from nasopharyngeal specimens or bronchoalveolar lavage specimens collected from children visiting Beijing Children's Hospital between 1997 and 2016. The hospital is a National Children's Medical Center, with more than 3 million outpatient visits and 70,000 hospitalizations every year. Among the 1865 isolates, 250 were serogroup 6, and the percentage rate was 13.4%. The distribution of the 250 serogroup 6 isolates by year was as follows: 1997, 34 isolates; 1998, 6 isolates; 1999, 6 isolates; 2000, 12 isolates; 2001, 8 isolates; 2002, 4 isolates; 2003, 6 isolates; 2004, 19 isolates; 2005, 11 isolates; 2008, 6 isolates; 2009, 10 isolates; 2010, 9 isolates; 2013, 41 isolates; 2014, 32 isolates; 2015, 23 isolates; 2016, 23 isolates. Because of the lack of personnel, the isolates collecting work was broken off in 2006–2007 and 2011–2012. All of the isolates were identified as serotype 6A, 6B, 6C, or 6D by Quellung test using antiserum provided by Statens Serum Institute (Copenhagen, Denmark).
The Ethics Committee of Beijing Children's Hospital affiliated to Capital Medical University exempted this study from review because it focused on the bacterial isolates and did not include patient data.
Antimicrobial susceptibility testingThe minimum inhibitory concentrations (MICs) of penicillin, amoxicillin-clavulanic acid, ceftriaxone, cefuroxime, erythromycin, imipenem, levofloxacin, and vancomycin for each of the isolates were determined using E-test strips (AB Biodisk, Solna, Sweden). Disc diffusion tests (Oxoid) were performed to ascertain antimicrobial susceptibilities to tetracycline, sulfamethoxazole-trimethoprim, and chloramphenicol. The results were interpreted in accordance with the Clinical and Laboratory Standards Institute 2016 guidelines, and both oral breakpoints (susceptible, ≤0.06mg/L; intermediate, 0.12–1mg/L; resistant, ≥2mg/L) and parenteral breakpoints (susceptible, ≤2mg/L; intermediate, 4mg/L; resistant, ≥8mg/L) were used for penicillin.17S. pneumoniae ATCC49619 was used as quality control.
Multilocus sequence typing analysisIsolates were characterized using multilocus sequence typing (MLST). Chromosomal DNA was extracted from overnight cultures of S. pneumoniae isolates grown on trypticase soy agar supplemented with 5% sheep blood (Oxoid Ltd, Basingstoke, England) using a SiMax Genomic DNA Extraction Kit (SBS Genetech Co., Ltd, China) according to the manufacturer's instructions. Seven housekeeping genes (aroE, gdh, gki, recP, spi, xpt, and ddl) were amplified via polymerase chain reaction from the chromosomal DNA as described previously.18 The products were sent to BGI (Beijing, China) for sequencing from both strands. The resulting sequences were compared with those of all known alleles at each of the loci, as well as with the sequence types (STs) recorded in the pneumococcal MLST database (https://pubmlst.org/spneumoniae/). New alleles and allelic profiles were submitted to the MLST database for name assignment. eBURST v3 software (available at http://eburst.mlst.net/) was used to evaluate the relationships among the isolates, and to assign strains to a clonal complex (CC) using the stringent group definition of six of seven shared alleles.19 STs sharing six identical alleles out of the seven MLST loci were assigned to the same CC.
Statistical analysisAntimicrobial susceptibility and MLST data were analyzed using WHONET 5.6 software as recommended by the World Health Organization (http://www.who.int/drugresistance/whonetsoftware/en/). The χ2 and/or Fisher's exact tests were used for analysis of categorical data, and statistical significance was set at p≤0.05.
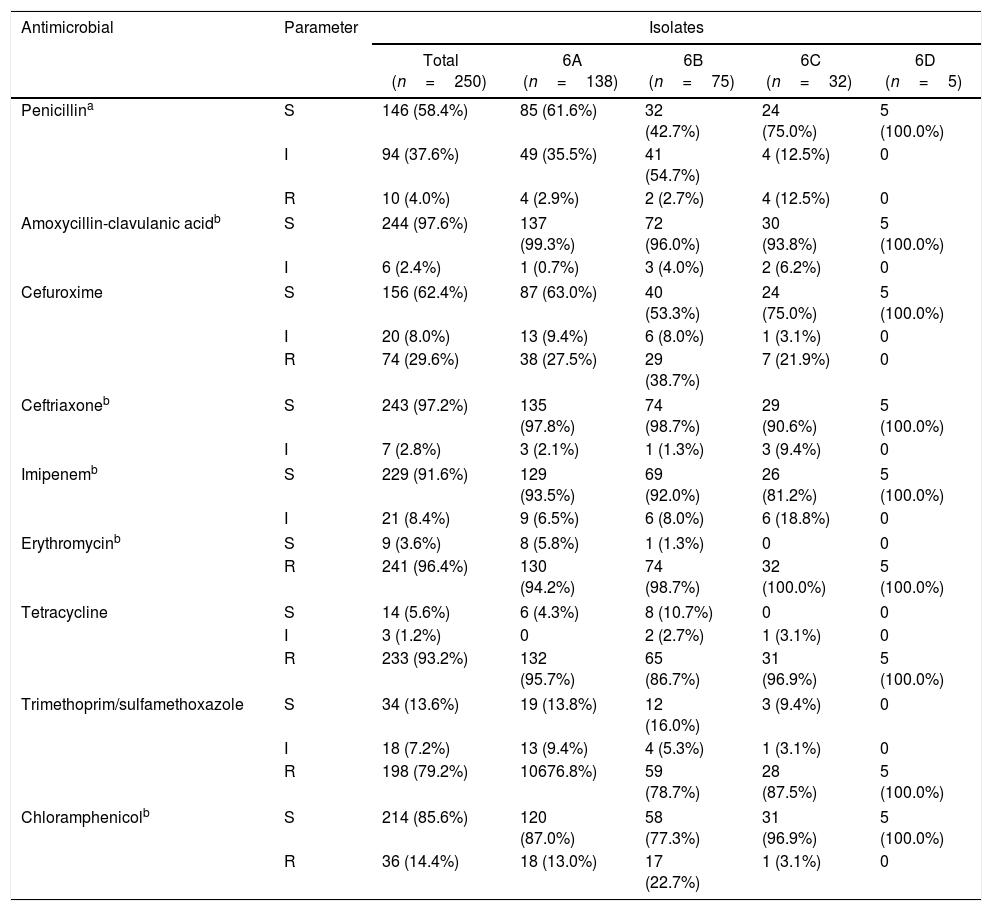
ResultsSerotyping and antimicrobial susceptibility testingAmong the 250 isolates, the rates of serotypes 6A, 6B, 6C, and 6D were 55.2% (138/250), 30.0% (75/250), 12.8% (32/250), and 2.0% (5/250), respectively. The results of antimicrobial susceptibility testing, including MIC values, for the different serotypes are shown in Table 1. All of the strains were susceptible to levofloxacin and vancomycin, and no resistance was observed toward amoxycillin-clavulanic acid, ceftriaxone, or imipenem. However, 96.4% (241/250) of isolates were resistant to erythromycin, with 236 isolates showing MIC values >256mg/L. None of the isolates were resistant to parenteral penicillin, but the rates of intermediate resistance and resistance to oral penicillin reached 37.6% (94/250) and 4.0% (10/250), respectively. Thus, penicillin resistance was defined according to the oral penicillin breakpoints. In addition, high rates of resistance to tetracycline (93.2%) were observed.
Susceptibility and minimum inhibitory concentration values of the Streptococcus pneumoniae isolates (grouped by serotype).
| Antimicrobial | Parameter | Isolates | ||||
|---|---|---|---|---|---|---|
| Total (n=250) | 6A (n=138) | 6B (n=75) | 6C (n=32) | 6D (n=5) | ||
| Penicillina | S | 146 (58.4%) | 85 (61.6%) | 32 (42.7%) | 24 (75.0%) | 5 (100.0%) |
| I | 94 (37.6%) | 49 (35.5%) | 41 (54.7%) | 4 (12.5%) | 0 | |
| R | 10 (4.0%) | 4 (2.9%) | 2 (2.7%) | 4 (12.5%) | 0 | |
| Amoxycillin-clavulanic acidb | S | 244 (97.6%) | 137 (99.3%) | 72 (96.0%) | 30 (93.8%) | 5 (100.0%) |
| I | 6 (2.4%) | 1 (0.7%) | 3 (4.0%) | 2 (6.2%) | 0 | |
| Cefuroxime | S | 156 (62.4%) | 87 (63.0%) | 40 (53.3%) | 24 (75.0%) | 5 (100.0%) |
| I | 20 (8.0%) | 13 (9.4%) | 6 (8.0%) | 1 (3.1%) | 0 | |
| R | 74 (29.6%) | 38 (27.5%) | 29 (38.7%) | 7 (21.9%) | 0 | |
| Ceftriaxoneb | S | 243 (97.2%) | 135 (97.8%) | 74 (98.7%) | 29 (90.6%) | 5 (100.0%) |
| I | 7 (2.8%) | 3 (2.1%) | 1 (1.3%) | 3 (9.4%) | 0 | |
| Imipenemb | S | 229 (91.6%) | 129 (93.5%) | 69 (92.0%) | 26 (81.2%) | 5 (100.0%) |
| I | 21 (8.4%) | 9 (6.5%) | 6 (8.0%) | 6 (18.8%) | 0 | |
| Erythromycinb | S | 9 (3.6%) | 8 (5.8%) | 1 (1.3%) | 0 | 0 |
| R | 241 (96.4%) | 130 (94.2%) | 74 (98.7%) | 32 (100.0%) | 5 (100.0%) | |
| Tetracycline | S | 14 (5.6%) | 6 (4.3%) | 8 (10.7%) | 0 | 0 |
| I | 3 (1.2%) | 0 | 2 (2.7%) | 1 (3.1%) | 0 | |
| R | 233 (93.2%) | 132 (95.7%) | 65 (86.7%) | 31 (96.9%) | 5 (100.0%) | |
| Trimethoprim/sulfamethoxazole | S | 34 (13.6%) | 19 (13.8%) | 12 (16.0%) | 3 (9.4%) | 0 |
| I | 18 (7.2%) | 13 (9.4%) | 4 (5.3%) | 1 (3.1%) | 0 | |
| R | 198 (79.2%) | 10676.8%) | 59 (78.7%) | 28 (87.5%) | 5 (100.0%) | |
| Chloramphenicolb | S | 214 (85.6%) | 120 (87.0%) | 58 (77.3%) | 31 (96.9%) | 5 (100.0%) |
| R | 36 (14.4%) | 18 (13.0%) | 17 (22.7%) | 1 (3.1%) | 0 | |
Note: All 250 isolates were susceptible to levofloxacin and vancomycin.
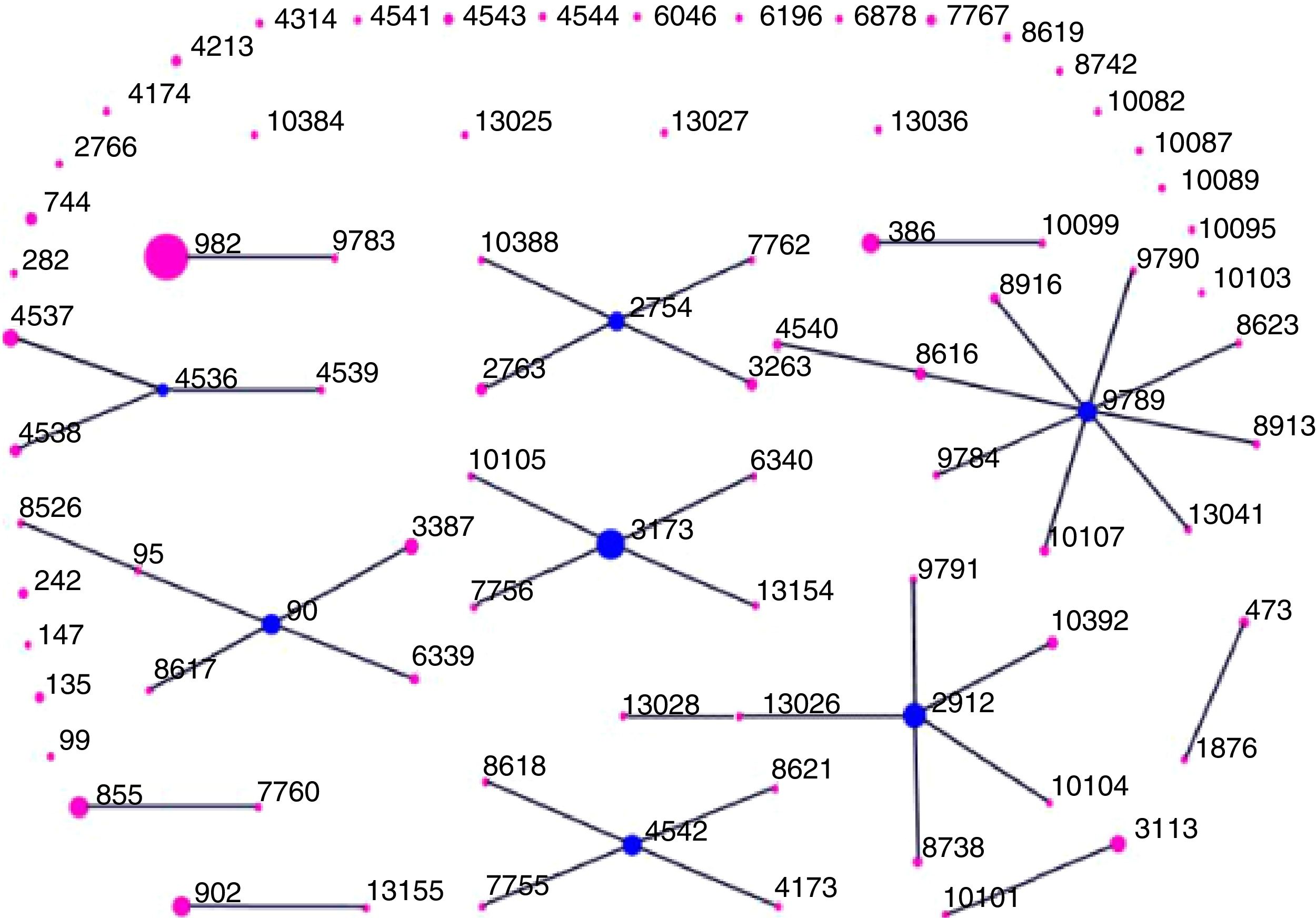
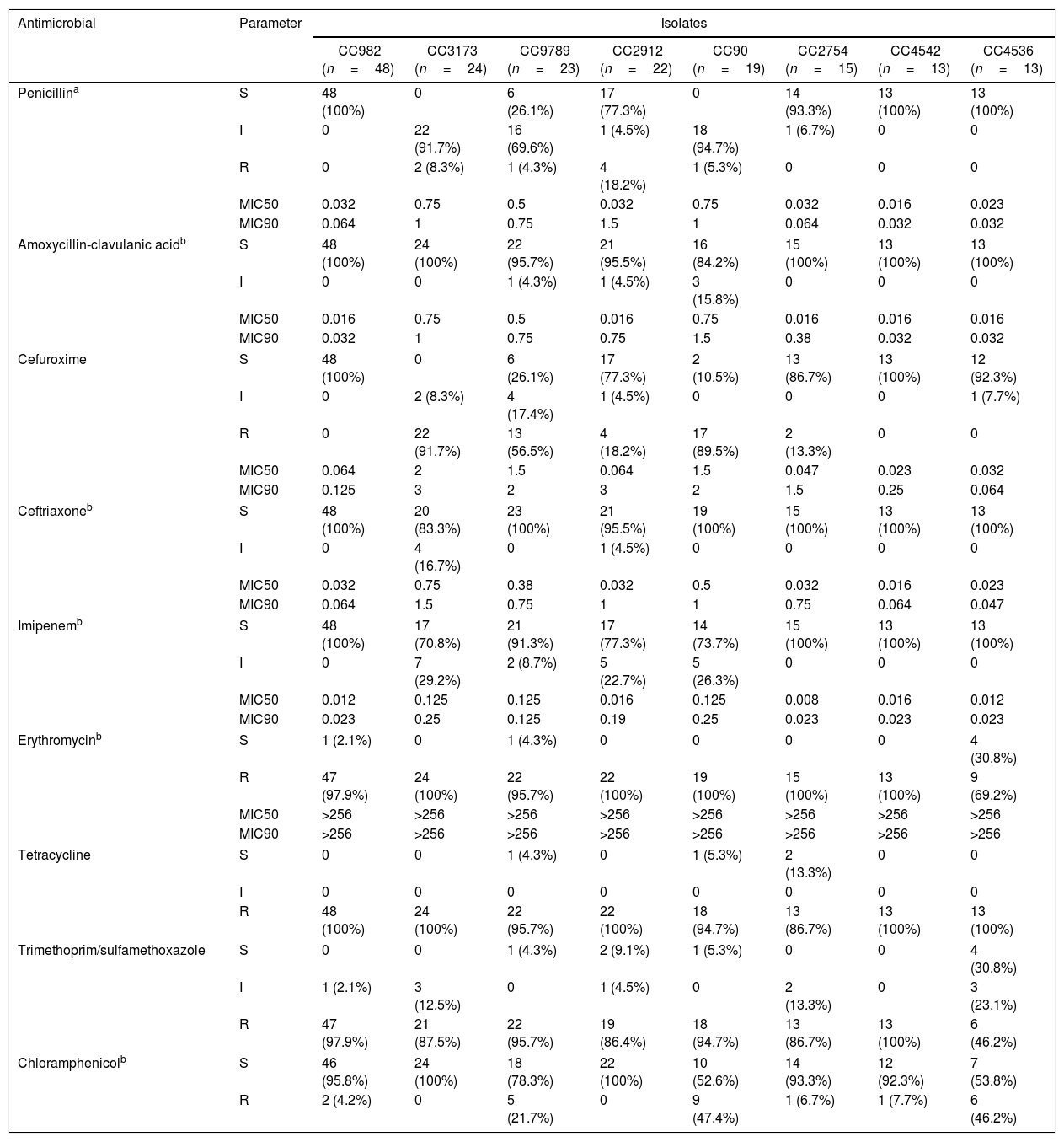
We identified 82 distinct STs among the 250 isolates, which were assigned using eBURST analysis to 13 CCs and 28 singletons (Fig. 1). CC982 was the predominant CC, accounting for 19.2% (48/250) of isolates, followed by CC3173 (9.6%, 24/250), CC9789 (9.2%, 23/250), CC2912 (8.8%, 22/250), CC90 (7.6%, 19/250), CC2754 (6.0%, 15/250), CC4542 (5.2%, 13/250), CC4536 (5.2%, 13/250), CC855 (4.0%, 10/250), CC902 (3.6%, 9/250), CC386 (3.6%, 9/250), CC3113 (2.8%, 7/250), and CC473 (1.2%, 3/250). Overall, these 13 CCs accounted for 86.0% (215/250) of the isolates.
Distribution of sequence types (STs) amongst the clonal complexes of Streptococcus pneumoniae. The image depicts eBURST analysis of the multilocus sequence typing data generated from all of the 250 Streptococcus pneumoniae isolates analyzed in this study. STs that are linked by a line belong to the same cluster. Circle sizes are proportional to the number of isolates within the ST.
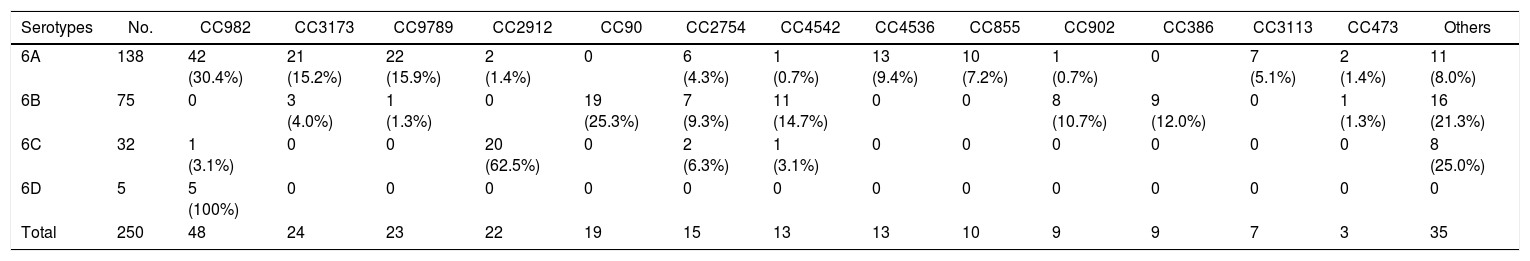
The distribution of CCs according to serotypes is shown in Table 2. Each serotype had a predominant CC that included the majority of the isolates. CC982 was the most prevalent CC amongst serotype 6A isolates, accounting for 30.4% (42/138) of isolates, followed by CC9789 (15.9%, 22/138) and CC3173 (15.2%, 21/138). For serotype 6B isolates, CC90 and CC4542 were the most common CCs, accounting for 25.3% (19/75) and 14.7% (11/75) of isolates, respectively. CC2912 accounted for 20 (62.5%) serotype 6C isolates, while all of the five serotype 6D isolates belonged to CC982. Interestingly, all of the CC4536, CC855, and CC3113 isolates were identified as serotype 6A, whereas CC90 and CC386 only contained serotype 6B isolates.
Distribution of clonal complexes/sequence types of serogroup 6 Streptococcus pneumoniae isolates by serotype.
| Serotypes | No. | CC982 | CC3173 | CC9789 | CC2912 | CC90 | CC2754 | CC4542 | CC4536 | CC855 | CC902 | CC386 | CC3113 | CC473 | Others |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 6A | 138 | 42 (30.4%) | 21 (15.2%) | 22 (15.9%) | 2 (1.4%) | 0 | 6 (4.3%) | 1 (0.7%) | 13 (9.4%) | 10 (7.2%) | 1 (0.7%) | 0 | 7 (5.1%) | 2 (1.4%) | 11 (8.0%) |
| 6B | 75 | 0 | 3 (4.0%) | 1 (1.3%) | 0 | 19 (25.3%) | 7 (9.3%) | 11 (14.7%) | 0 | 0 | 8 (10.7%) | 9 (12.0%) | 0 | 1 (1.3%) | 16 (21.3%) |
| 6C | 32 | 1 (3.1%) | 0 | 0 | 20 (62.5%) | 0 | 2 (6.3%) | 1 (3.1%) | 0 | 0 | 0 | 0 | 0 | 0 | 8 (25.0%) |
| 6D | 5 | 5 (100%) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Total | 250 | 48 | 24 | 23 | 22 | 19 | 15 | 13 | 13 | 10 | 9 | 9 | 7 | 3 | 35 |
%, Number of serotypes divided by number of clonal complexes/sequence types.
Table 3 summarizes the analysis of antibiotic resistance profiles according to CCs (Table 3). Antibiotic resistance profiles of isolates belonging to the same serotype varied according to the CC. Amongst the serotype 6A isolates, all of those belonging to CC982 and CC4536 were susceptible to penicillin, and the rates of susceptibility to cefuroxime were also high (CC982, 100%; CC4536, 92.3%). On the contrary, most of the CC3173, CC9789, and CC855 isolates were non-susceptible to penicillin and cefuroxime, with non-susceptibility rates reaching 100%, 73.9%, and 100% for penicillin and 100%, 73.9%, and 80.0% for cefuroxime, respectively.
Susceptibility and minimum inhibitory concentration values of the Streptococcus pneumoniae isolates grouped by clonal complex.
| Antimicrobial | Parameter | Isolates | |||||||
|---|---|---|---|---|---|---|---|---|---|
| CC982 (n=48) | CC3173 (n=24) | CC9789 (n=23) | CC2912 (n=22) | CC90 (n=19) | CC2754 (n=15) | CC4542 (n=13) | CC4536 (n=13) | ||
| Penicillina | S | 48 (100%) | 0 | 6 (26.1%) | 17 (77.3%) | 0 | 14 (93.3%) | 13 (100%) | 13 (100%) |
| I | 0 | 22 (91.7%) | 16 (69.6%) | 1 (4.5%) | 18 (94.7%) | 1 (6.7%) | 0 | 0 | |
| R | 0 | 2 (8.3%) | 1 (4.3%) | 4 (18.2%) | 1 (5.3%) | 0 | 0 | 0 | |
| MIC50 | 0.032 | 0.75 | 0.5 | 0.032 | 0.75 | 0.032 | 0.016 | 0.023 | |
| MIC90 | 0.064 | 1 | 0.75 | 1.5 | 1 | 0.064 | 0.032 | 0.032 | |
| Amoxycillin-clavulanic acidb | S | 48 (100%) | 24 (100%) | 22 (95.7%) | 21 (95.5%) | 16 (84.2%) | 15 (100%) | 13 (100%) | 13 (100%) |
| I | 0 | 0 | 1 (4.3%) | 1 (4.5%) | 3 (15.8%) | 0 | 0 | 0 | |
| MIC50 | 0.016 | 0.75 | 0.5 | 0.016 | 0.75 | 0.016 | 0.016 | 0.016 | |
| MIC90 | 0.032 | 1 | 0.75 | 0.75 | 1.5 | 0.38 | 0.032 | 0.032 | |
| Cefuroxime | S | 48 (100%) | 0 | 6 (26.1%) | 17 (77.3%) | 2 (10.5%) | 13 (86.7%) | 13 (100%) | 12 (92.3%) |
| I | 0 | 2 (8.3%) | 4 (17.4%) | 1 (4.5%) | 0 | 0 | 0 | 1 (7.7%) | |
| R | 0 | 22 (91.7%) | 13 (56.5%) | 4 (18.2%) | 17 (89.5%) | 2 (13.3%) | 0 | 0 | |
| MIC50 | 0.064 | 2 | 1.5 | 0.064 | 1.5 | 0.047 | 0.023 | 0.032 | |
| MIC90 | 0.125 | 3 | 2 | 3 | 2 | 1.5 | 0.25 | 0.064 | |
| Ceftriaxoneb | S | 48 (100%) | 20 (83.3%) | 23 (100%) | 21 (95.5%) | 19 (100%) | 15 (100%) | 13 (100%) | 13 (100%) |
| I | 0 | 4 (16.7%) | 0 | 1 (4.5%) | 0 | 0 | 0 | 0 | |
| MIC50 | 0.032 | 0.75 | 0.38 | 0.032 | 0.5 | 0.032 | 0.016 | 0.023 | |
| MIC90 | 0.064 | 1.5 | 0.75 | 1 | 1 | 0.75 | 0.064 | 0.047 | |
| Imipenemb | S | 48 (100%) | 17 (70.8%) | 21 (91.3%) | 17 (77.3%) | 14 (73.7%) | 15 (100%) | 13 (100%) | 13 (100%) |
| I | 0 | 7 (29.2%) | 2 (8.7%) | 5 (22.7%) | 5 (26.3%) | 0 | 0 | 0 | |
| MIC50 | 0.012 | 0.125 | 0.125 | 0.016 | 0.125 | 0.008 | 0.016 | 0.012 | |
| MIC90 | 0.023 | 0.25 | 0.125 | 0.19 | 0.25 | 0.023 | 0.023 | 0.023 | |
| Erythromycinb | S | 1 (2.1%) | 0 | 1 (4.3%) | 0 | 0 | 0 | 0 | 4 (30.8%) |
| R | 47 (97.9%) | 24 (100%) | 22 (95.7%) | 22 (100%) | 19 (100%) | 15 (100%) | 13 (100%) | 9 (69.2%) | |
| MIC50 | >256 | >256 | >256 | >256 | >256 | >256 | >256 | >256 | |
| MIC90 | >256 | >256 | >256 | >256 | >256 | >256 | >256 | >256 | |
| Tetracycline | S | 0 | 0 | 1 (4.3%) | 0 | 1 (5.3%) | 2 (13.3%) | 0 | 0 |
| I | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| R | 48 (100%) | 24 (100%) | 22 (95.7%) | 22 (100%) | 18 (94.7%) | 13 (86.7%) | 13 (100%) | 13 (100%) | |
| Trimethoprim/sulfamethoxazole | S | 0 | 0 | 1 (4.3%) | 2 (9.1%) | 1 (5.3%) | 0 | 0 | 4 (30.8%) |
| I | 1 (2.1%) | 3 (12.5%) | 0 | 1 (4.5%) | 0 | 2 (13.3%) | 0 | 3 (23.1%) | |
| R | 47 (97.9%) | 21 (87.5%) | 22 (95.7%) | 19 (86.4%) | 18 (94.7%) | 13 (86.7%) | 13 (100%) | 6 (46.2%) | |
| Chloramphenicolb | S | 46 (95.8%) | 24 (100%) | 18 (78.3%) | 22 (100%) | 10 (52.6%) | 14 (93.3%) | 12 (92.3%) | 7 (53.8%) |
| R | 2 (4.2%) | 0 | 5 (21.7%) | 0 | 9 (47.4%) | 1 (6.7%) | 1 (7.7%) | 6 (46.2%) | |
| Antimicrobial | Parameter | Isolates | ||||||
|---|---|---|---|---|---|---|---|---|
| CC855 (n=10) | CC902 (n=9) | CC386 (n=9) | CC3113 (n=7) | CC473 (n=3) | Others (n=35) | Total (n=250) | ||
| Penicillina | S | 0 | 0 | 8 (88.9%) | 7 (100%) | 0 | 20 (57.1%) | 146 (58.4%) |
| I | 10 (100%) | 8 (88.9%) | 1 (11.1%) | 0 | 3 (100%) | 14 (40.0%) | 94 (37.6%) | |
| R | 0 | 1 (11.1%) | 0 | 0 | 0 | 1 (2.9%) | 10 (4.0%) | |
| MIC50 | 0.25 | 0.5 | 0.064 | 0.023 | 0.25 | 0.064 | 0.047 | |
| MIC90 | 0.75 | 1.5 | 0.094 | 0.023 | 0.38 | 1 | 0.75 | |
| Amoxycillin-clavulanic acidb | S | 10 (100%) | 9 (100%) | 9 (100%) | 7 (100%) | 3 (100%) | 34 (97.1%) | 244 (97.6%) |
| I | 0 | 0 | 0 | 0 | 0 | 1 (2.9%) | 6 (2.4%) | |
| MIC50 | 0.5 | 0.38 | 0.064 | 0.016 | 0.125 | 0.032 | 0.032 | |
| MIC90 | 1 | 0.75 | 0.094 | 0.023 | 0.125 | 1 | 0.75 | |
| Cefuroxime | S | 2 (20.0%) | 0 | 9 (100%) | 7 (100%) | 3 (100%) | 24 (68.6%) | 156 (62.4%) |
| I | 5 (50.0%) | 5 (55.6%) | 0 | 0 | 0 | 2 (5.7%) | 20 (8.0%) | |
| R | 3 (30.0%) | 4 (44.4%) | 0 | 0 | 0 | 9 (25.7%) | 74 (29.6%) | |
| MIC50 | 1 | 1 | 0.094 | 0.094 | 0.38 | 0.094 | 0.094 | |
| MIC90 | 1.5 | 1.5 | 0.125 | 0.19 | 0.5 | 2 | 2 | |
| Ceftriaxoneb | S | 10 (100%) | 9 (100%) | 9 (100%) | 7 (100%) | 3 (100%) | 33 (94.3%) | 243 (97.2%) |
| I | 0 | 0 | 0 | 0 | 0 | 2 (5.7%) | 7 (2.8%) | |
| MIC50 | 0.38 | 0.5 | 0.064 | 0.125 | 0.125 | 0.047 | 0.064 | |
| MIC90 | 0.5 | 1 | 0.125 | 0.19 | 0.19 | 0.75 | 0.75 | |
| Imipenemb | S | 9 (90.0%) | 9 (100%) | 9 (100%) | 7 (100%) | 3 (100%) | 34 (97.1%) | 229 (91.6%) |
| I | 1 (10.0%) | 0 | 0 | 0 | 0 | 1 (2.9%) | 21 (8.4%) | |
| MIC50 | 0.094 | 0.094 | 0.032 | 0.008 | 0.032 | 0.023 | 0.023 | |
| MIC90 | 0.125 | 0.125 | 0.064 | 0.012 | 0.032 | 0.125 | 0.125 | |
| Erythromycinb | S | 0 | 0 | 0 | 0 | 0 | 3 (8.6%) | 9 (3.6%) |
| R | 10 (100%) | 9 (100%) | 9 (100%) | 7 (100%) | 3 (100%) | 32 (91.4%) | 241 (96.4%) | |
| MIC50 | >256 | >256 | >256 | >256 | >256 | >256 | >256 | |
| MIC90 | >256 | >256 | >256 | >256 | >256 | >256 | >256 | |
| Tetracycline | S | 0 | 2 (22.2%) | 0 | 0 | 2 (66.7%) | 6 (17.1%) | 14 (5.6%) |
| I | 0 | 1 (11.1%) | 0 | 0 | 0 | 2 (5.7%) | 3 (1.2%) | |
| R | 10 (100%) | 6 (66.7%) | 9 (100%) | 7 (100%) | 1 (33.3%) | 27 (77.1%) | 233 (93.2%) | |
| Trimethoprim/sulfamethoxazole | S | 9 (90.0%) | 0 | 8 (88.9%) | 0 | 3 (100%) | 6 (17.1%) | 34 (13.6%) |
| I | 1 (10.0%) | 1 (11.1%) | 0 | 3 (42.9%) | 0 | 3 (8.6%) | 18 (7.2%) | |
| R | 0 | 8 (88.9%) | 1 (11.1%) | 4 (57.1%) | 0 | 26 (74.3%) | 198 (79.2%) | |
| Chloramphenicolb | S | 9 (90.0%) | 9 (100%) | 9 (100%) | 7 (100%) | 2 (66.7%) | 25 (71.4%) | 214 (85.6%) |
| R | 1 (10.0%) | 0 | 0 | 0 | 1 (33.3%) | 10 (28.6%) | 36 (14.4%) | |
Note: All 250 isolates were susceptible to levofloxacin and vancomycin.
Of the serotype 6B isolates, all of the CC4542 isolates were susceptible to penicillin and cefuroxime, but CC902 isolates had a non-susceptibility rate of 100% for these two antibiotics. None of the CC90 isolates were susceptible to penicillin, and showed a cefuroxime resistance rate of 89.5%.
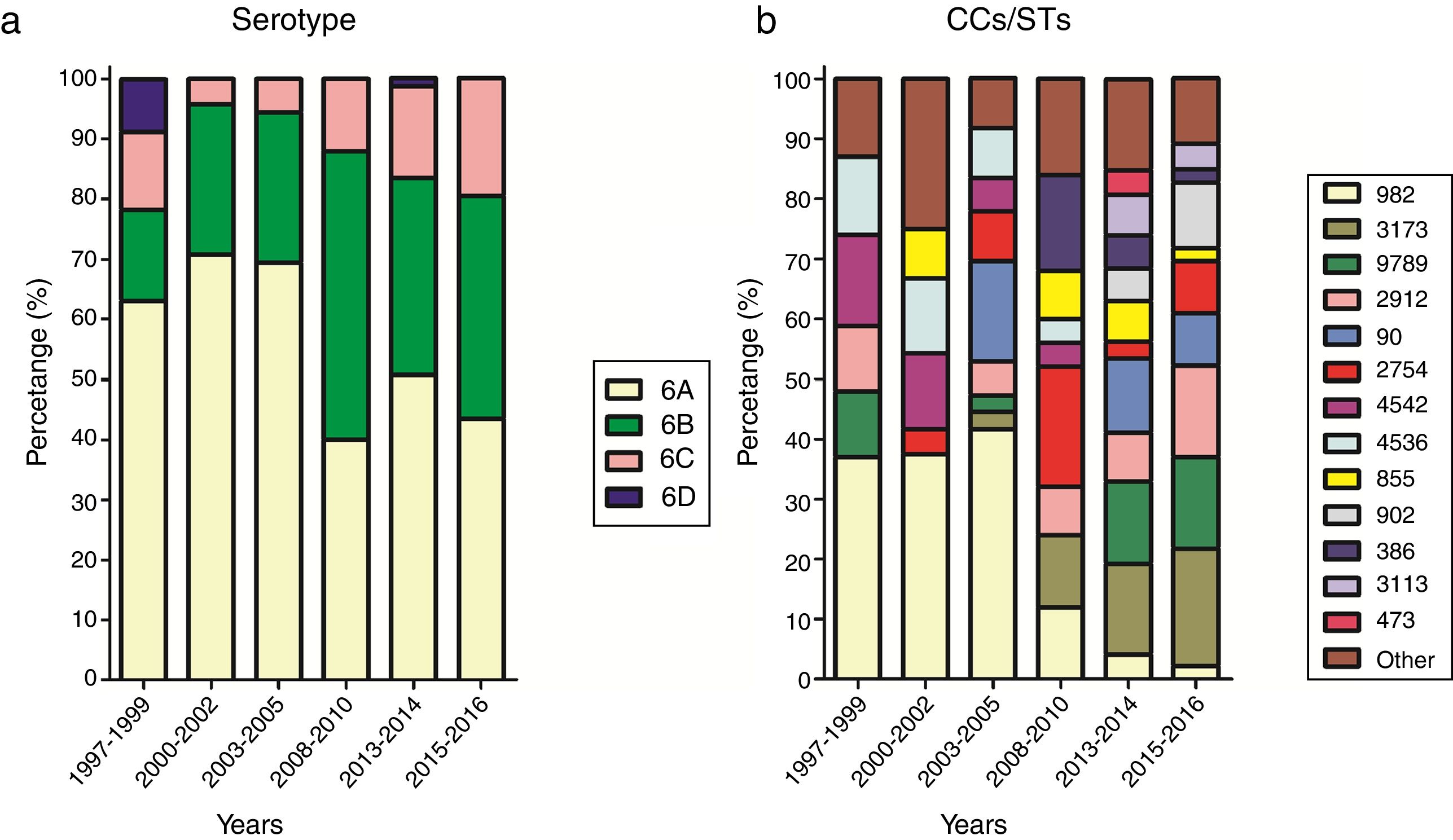
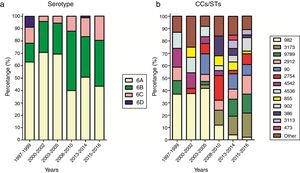
Fluctuation of serotypes and CCs between yearsFor effective comparison, the study period was divided into six ranges: 1997–1999, 2000–2002, 2003–2005, 2008–2010, 2013–2014, and 2015–2016. The distribution of serotypes and CCs in the different periods is shown in Fig. 2. A steady increase in the prevalence of serotype 6C could be seen from 2008, while the prevalence of serotype 6A, which was the most common serotype, decreased over the study period (χ2=11.963, p<0.05).
Over the study period, the percentage of CC982 isolates decreased from 37.0% in 1997–1999 to 2.2% in 2015–2016. The percentage of CC982, CC4542, and CC4536 isolates showing high susceptibility to penicillin and cefuroxime decreased over the study period, with no susceptible isolates identified after 2010, while the proportion of CC3173, CC9789, CC855, and CC902 isolates showing non-susceptibility to penicillin and cefuroxime increased.
DiscussionThis study investigated the antibiotic susceptibility profiles and fluctuations in the dominant CCs of serogroup 6 S. pneumoniae strains isolated from children in Beijing, the capital city of China, between 1997 and 2016. Of the 250 isolates examined, 55.2%, 30.0%, 12.8%, and 2.0% were identified as serotypes 6A, 6B, 6C, and 6D, respectively. Although the majority of the isolates belonged to serotype 6A, the prevalence of this serotype decreased over the study period. Research from countries with high rates of PCV7 vaccination have shown serotype replacement of serogroup 6. Significant decreases in the prevalence of serotype 6A and 6B strains were observed in the UK between 2006/2007 and 2010/2011, coinciding with the addition of PCV7 to the UK National Immunization Program in September 2006, and its replacement with PCV13 in April 2010.11 A study from Israel conducted from 1999 to 2014, pre- and post-PCV implementation, analyzed the impact of PCVs on the prevalence of serogroup 6 carriage, and the incidence of pneumococcal conjunctivitis, otitis media, and invasive diseases caused by serogroup 6. The researchers found that the carriage rates of serotype 6A and 6B isolates, along with cases of conjunctivitis, otitis media, and invasive diseases caused by serotype 6A and 6B, decreased remarkably in the post-PCV7 period. However, this decrease was offset by a significant increase in the prevalence of serotype 6C isolates, although this was halted after the implementation of PCV13.20 Several other studies report a similar phenomenon, whereby the prevalence of 6A and 6B isolates decreased with the introduction of PCV7 or PCV13, but the proportion of 6C isolates increased until the introduction of PCV13.21,22 The efficacy of PCV13 against serotype 6C isolates results from the extensive structural and immunological similarity between serotypes 6A and 6C. The inclusion of 6A in PCV13 results in a higher concentration of anti-6C antibodies than those obtained by PCV7.23 We also detected an increase of serotype 6C in the current study, although there were not many 6C isolates overall.
S. pneumoniae MLST is based on sequence data from standardized fragments of seven housekeeping genes. The MLST classification reveals important insights into the geographic spread of successful pathogenic clones, as well as the emergence of associations between STs and serotypes, combinations that can be traced back to serotype switching events. Among S. pneumoniae isolates, antimicrobial resistance associated with specific pneumococcal serotypes, even CCs/STs, most often reflects the spread of a few internationally disseminated clones recognized by the Pneumococcal Molecular Epidemiology Network (PMEN; http://www.pneumogen.net/pmen/). These clones are frequently associated with disease, widely distributed geographically, and usually resistant to one or more clinically relevant antibiotics.24
Data from the current study showed fluctuations in the dominance of particular CCs. The proportion of CCs with high rates of non-susceptibility to penicillin and cefuroxime, such as CC3173, CC9789, CC855, and CC902, increased during the study period. On the contrary, CC982, CC4542, and CC4536, all of which show high rates of susceptibility to penicillin and cefuroxime, decreased or even disappeared. Previous studies have also shown fluctuation of CCs/STs. For example, researches from United States,25 Canada26 and Russia27 showed that amongst serotype 19A isolates, CC320, which has a multidrug-resistant phenotype with high β-lactam MICs, replaced CC230, which is associated with high susceptibility to penicillin. Our previous researches from Beijing, China indicated that the globally-disseminated resistant clone CC271, which has a high rate of non-susceptibility to β-lactam antibiotics, took the place of CC983 amongst serotype 19F pneumococcus strains,28 while amongst serotype 23F isolates, ST342 was replaced by ST81, which demonstrates higher β-lactam resistance.29 Also our study on serotype 14 strains from Beijing, China reveled that CC876 overtook CC875 as the dominant CC, with CC876 isolates showing higher non-susceptibility rates to β-lactam antibiotics than CC875 isolates.30 All of these examples of clonal shift phenomena were assumed to be caused by antibiotic pressure, as CCs/STs with high rates of antibiotic resistance replaced those with low resistance.
Taken together, these observations underline the importance of further long-term surveillance of S. pneumoniae based on serotype and genotype, combined with antimicrobial susceptibility profile analysis, in the hopes of preventing infections caused by this important human pathogen.
Ethical statementThe pneumococcal isolates involved in this study were collected previously during a series of epidemiological surveys on this bacterium. The Ethics Committee of Beijing Children's Hospital affiliated to Capital Medical University exempted this study from review because it focused on the bacterial isolates and did not include patient data.
Authors’ contributionsAll of the authors participated in the design of the study. WS, YL, QM, LY, and WG collected the isolates and performed the antimicrobial susceptibility test. WS, YL, and QM did the MLST part. WS, YL and KY collected the data, analyzed them, interpreted the results, and drafted the manuscript. All authors reviewed and revised the manuscript and approved the final version.
FundingThis study was supported by the Research Funds of Profession Quota Budget from Beijing Municipal Science and Technology Commission [grant number 2016-bjsekyjs-3] and the Beijing Talents Fund [grant number 2015000021469G209]. The sponsors had no role in study design, in the collection, analysis, and interpretation of data, in the writing of the report, or in the decision to submit the article for publication.
Conflicts of interestNone declared.
We thank Tamsin Sheen, PhD, from Liwen Bianji, Edanz Editing China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.