Matrix Assisted Laser Desorption/Ionization and Time of Flight mass spectrometry (MALDI-TOF MS) is a powerful tool for the identification of bacteria through the detection and analysis of their proteins or fragments derived from ribosomes. Slight sequence variations in conserved ribosomal proteins distinguish microorganisms at the subspecies and strain levels. Characterization of Leptospira spp. by 16S RNA sequencing is costly and time-consuming, and recent studies have shown that closely related species (e.g., Leptospira interrogans and Leptospira kirschneri) may not be discriminated using this technology. Herein, we report an in-house Leptospira reference spectra database using Leptospira reference strains that were validated with a collection of well-identified Brazilian isolates kept in the Bacterial Zoonosis Laboratory at the Veterinary Preventive Medicine and Animal Health Department at Sao Paulo University. In addition, L. interrogans and L. kirschneri were differentiated using an in-depth mass spectrometry analysis with ClinProTools™ software. In conclusion, our in-house reference spectra database has the necessary accuracy to differentiate pathogenic and non-pathogenic species and to distinguish L. interrogans and L. kirschneri.
Leptospirosis is a mammalian zoonosis caused by Leptospira strains belonging to the order Spirochaetales. Mammals, including humans, are affected by different clinical manifestations, depending on the virulence, motility, and ability of the leptospiral pathogen to survive in the host. Susceptibility to infection is dependent on age, genetic factors and skin integrity during the infection. Leptospira biology and leptospirosis physiopathology were comprehensively presented and discussed in a recent publication.1 The antigenic diversity among serovars differentiates pathogenic (Leptospira interrogans) and non-pathogenic or saprophytic (Leptospira biflexa) species.2 At least 22 species have been classified by molecular techniques.2–4 The microscopic agglutination test (MAT) is the most commonly used diagnostic method in the clinic; however, limitations have been previously reported and discussed.3,5 The characterization of Leptospira spp. using molecular techniques such as 16S RNA sequencing is costly and time-consuming,6 especially taking into account the large number of microorganisms identified in the clinical practice. This method depends on one or several target genes, however the data for all the peptides with a mass range of 2–20kDa could be collected using MALDI-TOF MS as demonstrated by Xiao et al.7 on molecular fingerprinting of pathogenic and non-pathogenic Leptospira. MALDI-TOF MS is a well-established technique for the rapid characterization of bacteria, and its use is continuously increasing.8 This technology can differentiate microorganisms’ species by the analysis and comparison of proteins or protein fragments derived from ribosomes. It is important to note that slight sequence variations in conserved ribosomal proteins are sufficient to distinguish microorganisms at the subspecies and strain levels.8 MALDI-TOF MS has been proposed to be a powerful tool for the identification of Leptospira at the species level.6–8,10 However, the misidentification of L. interrogans as L. kirschneri by MALDI-TOF MS has been described, and potential biomarkers to differentiate these species have been investigated.10 In the present paper, we focused on (i) the characterization of pathogenic and non-pathogenic Leptospira species of a Leptospira Brazilian collection using MALDI-TOF MS after creating an in-house database and (ii) the differentiation of L. interrogans from L. kirschneri by in-depth mass spectrometry analysis.
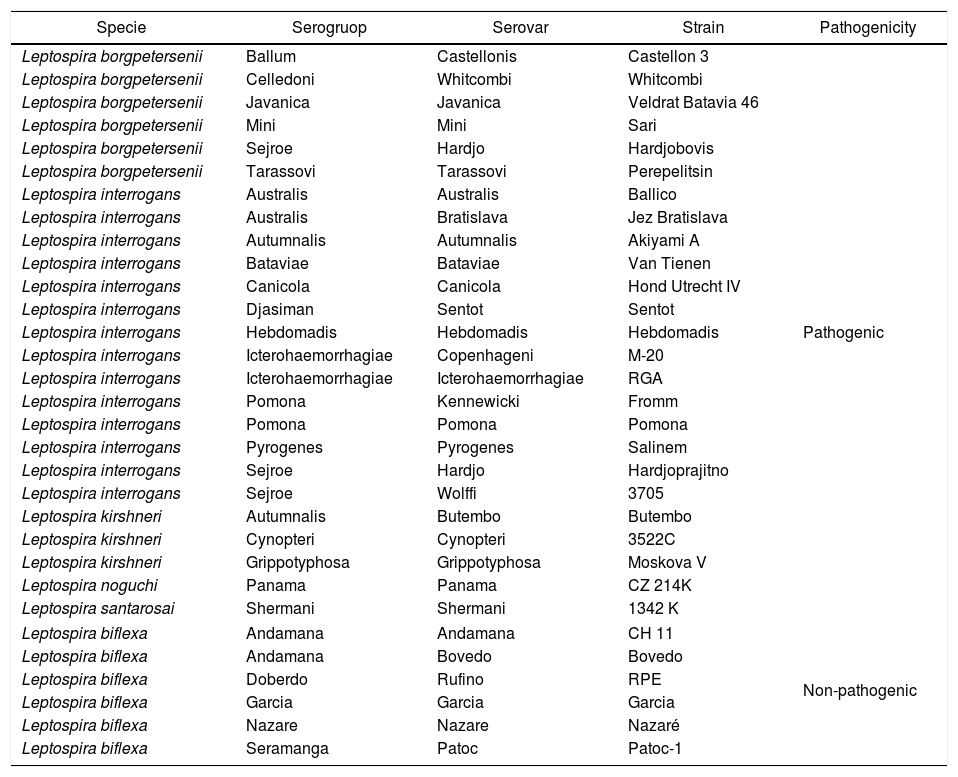
Material and methodsLeptospira strains and isolatesThirty-one reference leptospiral strains and 22 field isolates belonging to pathogenic (Leptospira interrogans, Leptospira borgpetersenii, Leptospira kirschneri, Leptospira noguchii and Leptospira santarosai) and non-pathogenic (Leptospira biflexa) species were analyzed. The Leptospira isolates were recovered from bovine, dog, human, Rattus norvegicus, and Rattus rattus urine samples taken from Sao Paulo, Rio de Janeiro and Londrina (Table 1). The strains and isolates were maintained in the Laboratory of Bacterial Zoonosis – School of Veterinary Medicine and Animal Science/University of Sao Paulo (USP) and stored in Fletcher semi-solid medium (Fletcher Medium Base, Difco™, NJ, USA) at 30°C. The species of the field isolates were previously identified by 16S rRNA sequencing (data not shown).
Leptospira strains used as reference for MALDI-TOF MS measurements.
| Specie | Serogruop | Serovar | Strain | Pathogenicity |
|---|---|---|---|---|
| Leptospira borgpetersenii | Ballum | Castellonis | Castellon 3 | Pathogenic |
| Leptospira borgpetersenii | Celledoni | Whitcombi | Whitcombi | |
| Leptospira borgpetersenii | Javanica | Javanica | Veldrat Batavia 46 | |
| Leptospira borgpetersenii | Mini | Mini | Sari | |
| Leptospira borgpetersenii | Sejroe | Hardjo | Hardjobovis | |
| Leptospira borgpetersenii | Tarassovi | Tarassovi | Perepelitsin | |
| Leptospira interrogans | Australis | Australis | Ballico | |
| Leptospira interrogans | Australis | Bratislava | Jez Bratislava | |
| Leptospira interrogans | Autumnalis | Autumnalis | Akiyami A | |
| Leptospira interrogans | Bataviae | Bataviae | Van Tienen | |
| Leptospira interrogans | Canicola | Canicola | Hond Utrecht IV | |
| Leptospira interrogans | Djasiman | Sentot | Sentot | |
| Leptospira interrogans | Hebdomadis | Hebdomadis | Hebdomadis | |
| Leptospira interrogans | Icterohaemorrhagiae | Copenhageni | M-20 | |
| Leptospira interrogans | Icterohaemorrhagiae | Icterohaemorrhagiae | RGA | |
| Leptospira interrogans | Pomona | Kennewicki | Fromm | |
| Leptospira interrogans | Pomona | Pomona | Pomona | |
| Leptospira interrogans | Pyrogenes | Pyrogenes | Salinem | |
| Leptospira interrogans | Sejroe | Hardjo | Hardjoprajitno | |
| Leptospira interrogans | Sejroe | Wolffi | 3705 | |
| Leptospira kirshneri | Autumnalis | Butembo | Butembo | |
| Leptospira kirshneri | Cynopteri | Cynopteri | 3522C | |
| Leptospira kirshneri | Grippotyphosa | Grippotyphosa | Moskova V | |
| Leptospira noguchi | Panama | Panama | CZ 214K | |
| Leptospira santarosai | Shermani | Shermani | 1342 K | |
| Leptospira biflexa | Andamana | Andamana | CH 11 | Non-pathogenic |
| Leptospira biflexa | Andamana | Bovedo | Bovedo | |
| Leptospira biflexa | Doberdo | Rufino | RPE | |
| Leptospira biflexa | Garcia | Garcia | Garcia | |
| Leptospira biflexa | Nazare | Nazare | Nazaré | |
| Leptospira biflexa | Seramanga | Patoc | Patoc-1 | |
Collection at the Bacterial Zoonoses Laboratory, Department of Veterinary Preventive and Animal Health of School of Veterinary Medicine and Animal Science, São Paulo University, Brazil.
The strains and isolates were grown and diluted (1:25) for seven days at 30°C in Ellinghausen-McCullough-Johnson-Harris medium (EMJH Difco™, NJ, USA), and bacterial cells were counted using a Petroff Hausser counting chamber (HS Hausser Scientific, Horsham, PA) by dark field microscopy. Leptospira cultures were centrifuged at 11,000×g for 10min at room temperature, and the pellet was washed twice with 3mL of phosphate buffered saline (PBS) and suspended in sterile deionized water to a final bacterial concentration of 1×108 organisms per mL. Ethanol/formic acid protein extraction was performed by addition of 300μL of the culture into 900μL of ethanol (99.8%, PA) followed vortexing and 10-min of incubation. This inactivation procedure was followed by a 10-min centrifugation at 11,000×g at room temperature, the supernatant was removed, and the pellet was air dried until the ethanol was completely evaporated. This process was repeated and the material was then dissolved in 30μL of 70% formic acid (Sigma–Aldrich) followed by the addition of 30μL of acetonitrile (Fluka Analytical Sigma–Aldrich, Munich, Germany). Centrifugation was performed at 11,000×g for 2min at room temperature. Two microliters of the clear supernatant were spotted on a 384 target polished steel plate (Bruker Daltonik GmbH, Bremen, Germany) and allowed to dry. Following this, the dried spot was overlaid with 2μL of matrix solution, a saturated solution of α-Cyano-4-hydroxycinnamic acid (HCCA, 99% Bruker Daltonik GmbH, Bremen or Sigma–Aldrich, Munich, Germany) (10mg/mL) in acetonitrile (Fluka Analytical Sigma–Aldrich) and 0.1% trifluoracetic acid (1:2) (TFA-Reagent PlusW 99%, Sigma–Aldrich). Finally, samples were allowed to dry at room temperature. Escherichia coli DH5α was used as a positive control, and a non-inoculated matrix solution was used as a negative control. During data acquisition, it was observed that some isolates underwent osmotic lysis in deionized water, which was corrected by replacing sterile deionized water by saline solution (0.85% NaCl) buffered with Sorensen's solution (69mM Na2HPO4/8mM NaH2PO4, pH 7.6).9 This solution has lower osmolarity than PBS, but kept cells intact without interfering with the ionization of the bacterial proteins as well as the mass fingerprint of our previously data that were generated in saline solution. Additional mass spectra were then obtained with fresh culture passages to ensure the minimum number of spectra for the generation of single Main Spectrum Profiles (MSP).
Instrument settings for MALDI-TOF MS analysisA Microflex LT™ (Bruker Daltonics, Bremen, Germany) instrument was used with the software Flex Control™ version 3.4 (Bruker Daltonics). For mass calibration and instrument parameter optimization bacterial test standard (BTS, Bruker Daltonics) was used. The acquisitions were done in linear positive mode within a mass range from 2000 to 20,000m/z with the manufacturer's suggested settings in automated collecting spectra mode.
All spectra were analyzed by standard pattern-matching algorithm using the MALDI Biotyper™ 3.1 software (Bruker Daltonics), and the raw spectra were compared with the reference spectra of the Bruker library (database version 3.3.1, 5627 reference spectra) with default settings. The ID criteria used was the recommended by the manufacturer: – a score ≥2.000 indicated species, a score between 1.700 and 1.999 indicated genus level and a score <1.700 was interpreted as no ID. For MainSpectra (MSP) and dendrogram construction, flat-liners and bad quality spectra were removed and additional measurements were carried out to obtain 20 spectra from each isolate/strain. Spectra were then loaded into Biotyper™ 3.1 software (Bruker Daltonics) for MSP creation and dendrogram clustering construction with the default settings (distance measure: correlation; linkage: average; score oriented). Each spot was measured in 1000-shot steps for a total of 4000 laser shots. Preparation of the BTS and calibration were performed following the manufacturer's instructions: calibration was successful when proteins of the mass spectra were in a range of ±200 parts per million (ppm).
In-house database and dendrogram constructionFor each of the 31 strains, 30 individual spectra were used to create a MSP. Flat-liners and bad quality spectra were removed, and additional measurements were carried out to obtain 30 spectra from each isolate/strain. The MSP was obtained using MALDI-Biotyper software (Bruker Daltonics, Germany) and then loaded into the Bruker Daltonics database (version 3.1.2.0). Software settings for MSP creation were set to maximal mass error of each single spectrum: 2000; desired mass error for the MSP: 200; desired peak frequency minimum (%): 25; and maximal desired peak number of the MSP: 70. Dendrogram clustering was constructed with the default setting of 160 (distance measure: correlation; linkage: average; score oriented).
Determining the efficiency of the database search with Leptospira field isolatesFour mass spectra of each field isolate were obtained and challenged against our in-house Leptospira database. The results were expressed in log score values, with values ≥2 indicating reliable species identification and values from 1.7 to 2.00 indicating reliable genus identification.
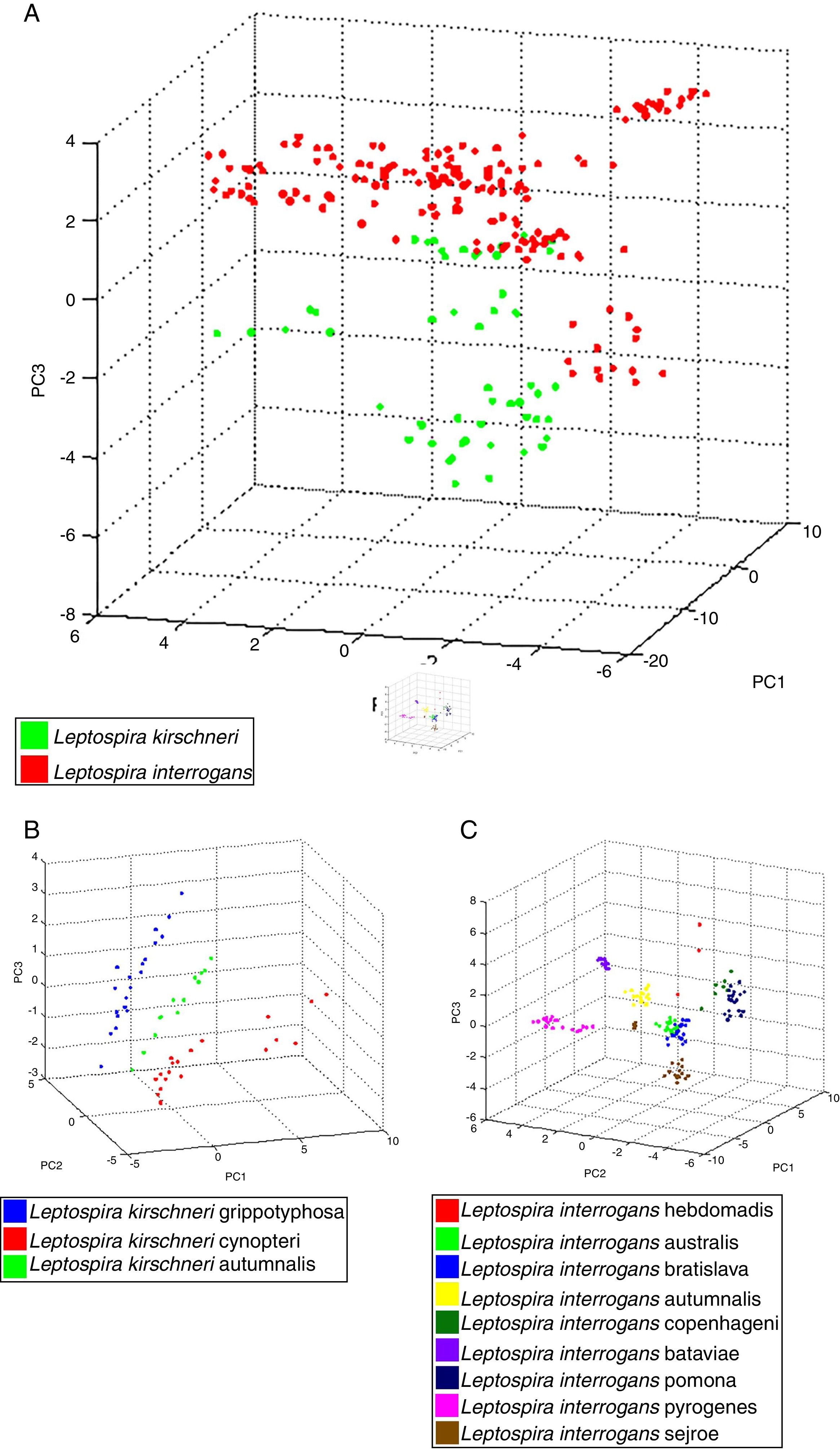
Differentiation of Leptospira interrogans and Leptospira kirschneri using ClinProTools™ClinProTools™ (Bruker Daltonics) generates multiple mathematical algorithms to generate pattern recognition models for the classification and prediction of different classes (e.g., L. interrogans class 1, L. kirschneri class 2) from mass spectrometry-based profiling data. Various spectra of the different serovars (03 serovar to L. kirschneri and 12 serovar to L. interrogans) were used for each class, seeking to standardize the data for species distinction. Moreover, ClinProTools™ provides a list of peaks sorted according to the statistical significance to differentiate between both classes.12 Thus, to recognize mass spectra patterns and biomarkers between L. interrogans and L. kirschneri, spectra peak analysis models with ClinProTools™ software v.3.0 (Bruker Daltonics) were created from an additional 210 mass spectra of 11 L. interrogans (15 high-quality mass spectra per isolate) and 3 L. kirschneri (15 high-quality mass spectra per isolate) isolates. Spectra were pretreated with a resolution of 800ppm, a mass range of 2000–20,000Da, a top hat baseline subtraction with 10% minimal baseline width, enabling null spectra exclusion, recalibration with 500ppm maximal peak shift and 30% match celebrant peaks. ClinProTools™ models (Bruker Daltonics) were generated using three algorithms: Genetic Algorithm (GA), Supervised Neural Network (SNN), and Quick Classifier (QC). For each model, the recognition capability (RC) and cross validation (CV) percentage were generated to demonstrate the reliability and accuracy of the model. RC and CV percentages are indicators of the model's performance and serve as useful predictors of the model's ability to classify test isolates. We also carried out principal component analysis (PCA) included in ClinProTools software aiming to visualize homogeneity and heterogeneity of the protein spectra of L. interrogans and L. kirschneri. Principal component analysis (PCA) and the results were shown in 3D score plot.
Single-peak analysisFor each peak, the AUC for the discrimination of the groups was directly obtained from the ClinProTools™ v.3.0 software (Bruker Daltonics). For the five peaks with the highest AUC, the detection performances were verified using FlexAnalysis™ v.3.4 (Bruker Daltonics). After smoothing and baseline subtraction, the mass lists for each isolate were obtained using the centroid algorithm with a signal-to-noise (SN) threshold of 0.5 and a maximum of 500 peaks and exported to Microsoft Excel. The SN ratios of the peaks with a tolerance of 1000ppm were exported to SPSS version 18.0. ROC (Receiver Operating Characteristic) curves were constructed, and their optimal cutoff values were determined with the maximum Youden index.11
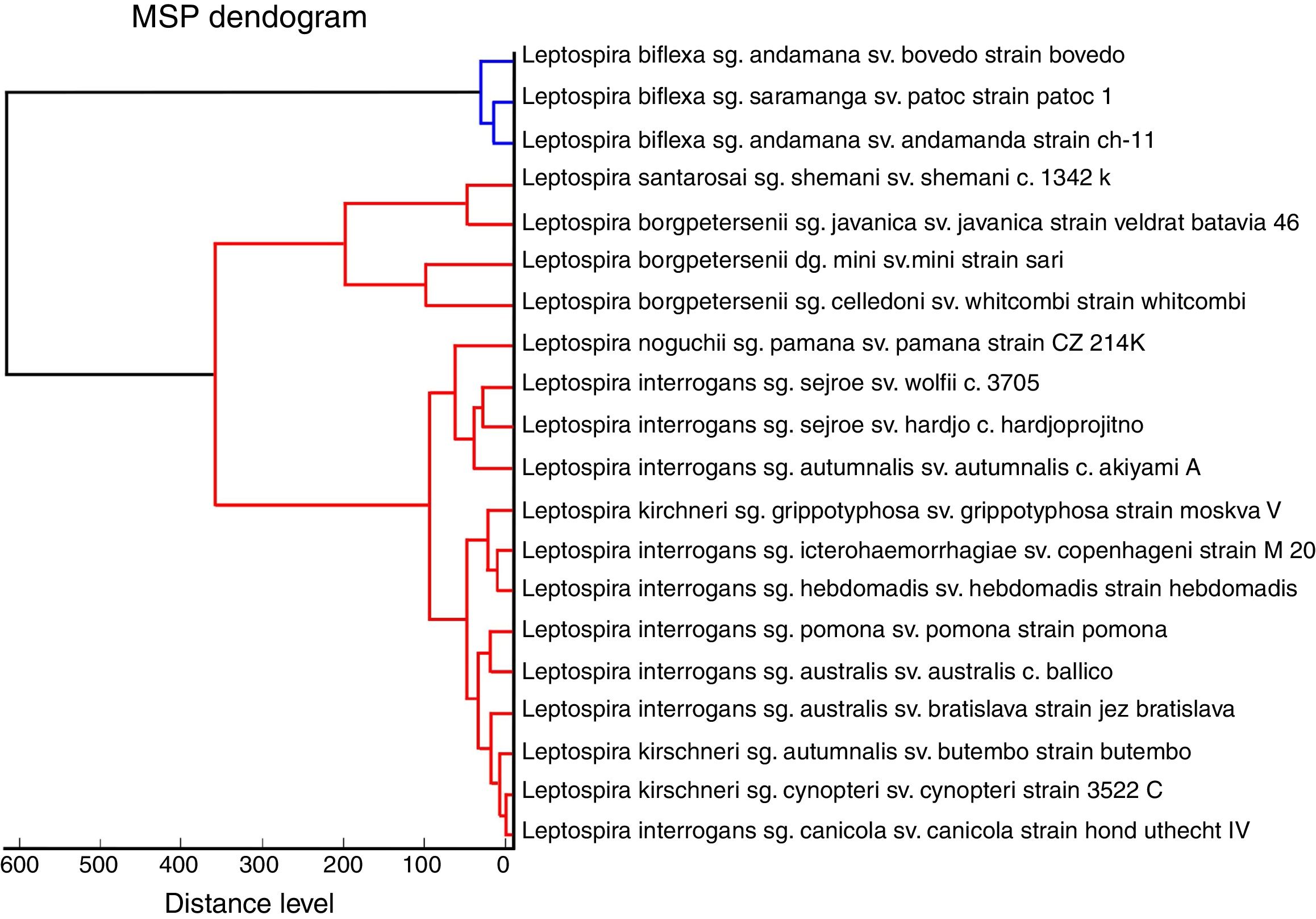
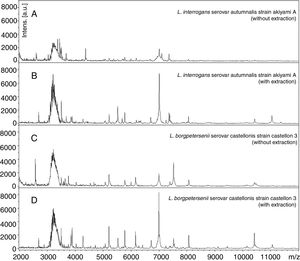
ResultsReference spectra were created for all 31 leptospiral strains and applied as unassigned MSPs in the commercially available MALDI Biotyper™ database spectra library, which lacks leptospiral protein profiles (Fig. 1). The MSPs were clustered according to pathogenicity in the MALDI-TOF MS dendrogram, and the pathogenic species (red) are clearly differentiated from the nonpathogenic Leptospira species (green) (Fig. 2). Similarly, the pathogenic L. borgpetersenii and L. interrogans are located in separate clusters, but, as expected, poor discrimination was obtained for L. interrogans and L. kirschneri.
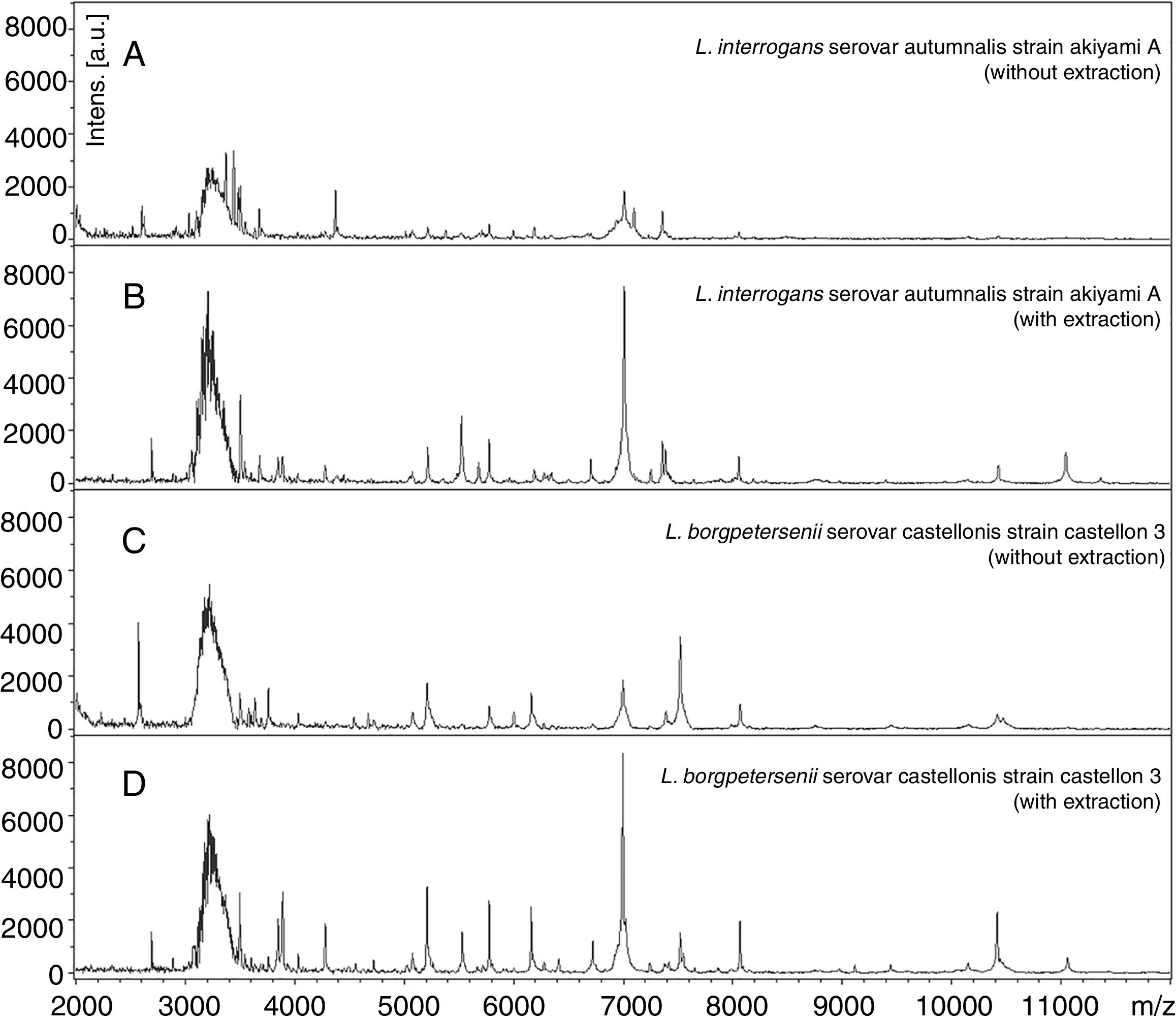
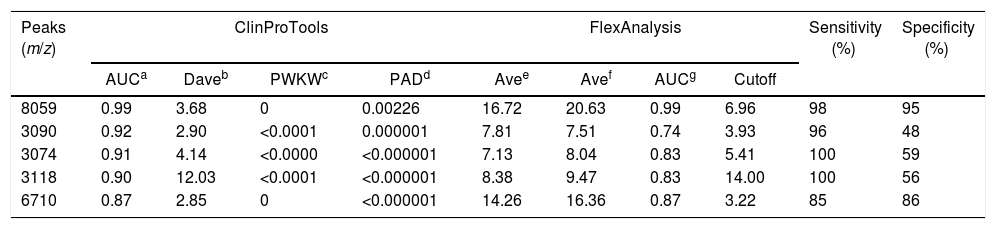
MALDI-TOF MS spectra obtained by analyzing the reference strains of Leptospira interrogans and Leptospira borgpetersenii with and without extraction as described in “Material and methods” section. These data show the importance of the protein extraction to obtain the better quality of spectra.
Representative mass spectra of L. interrogans and L. borgpetersenii obtained by direct analysis and by protein extraction protocol are shown in Fig. 1. In A and C, mass spectra were obtained without protein extraction and peaks with low intensity were observed. In contrast, B and D show higher quality mass spectra obtained after protein extraction protocol, with peaks with higher intensity.
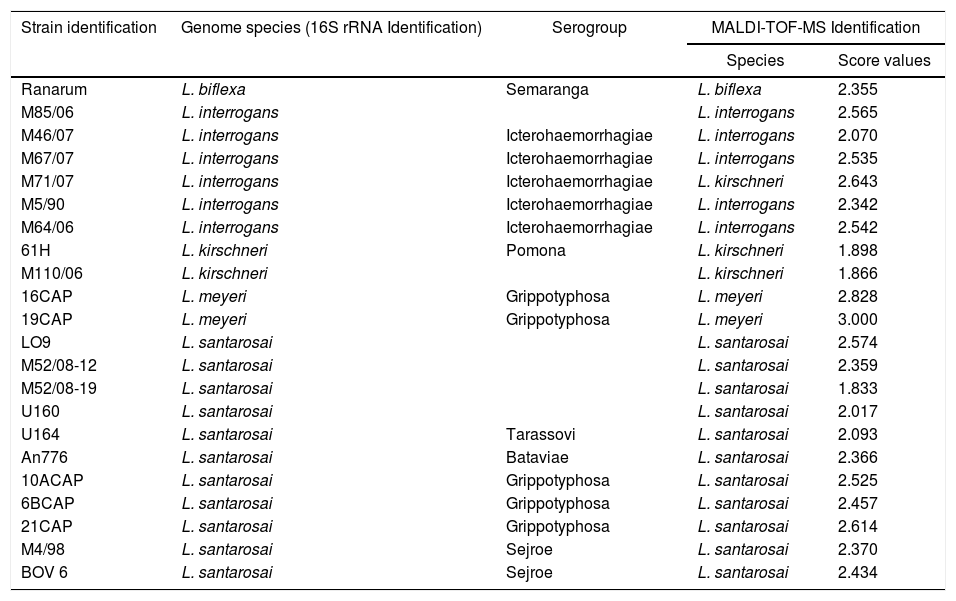
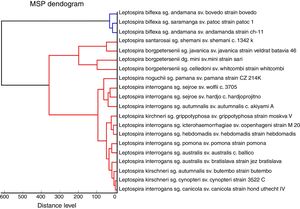
The 22 field isolates belonging to L. biflexa, L. interrogans and L. santarosai had the correct species assigned by MALDI-TOF MS, and all isolates showed score values over 2.0 (Table 2), where it is possible to identify all isolates by the correct species ID following our in-house database. The PCA reproduces through different statistical tests the reduction of several variables of a set of data, where each point represents a spectrum and each color represents a grouping of similar data. Fig. 3A presents the PCA for L. interrogans species in red and L. kirschneri in green, there is a perceptible distinction between the two groups even with some closer points showing that the PCA analysis does not guarantee a clear separation between the species. B presents the PCA for the serovars that formed the class L. kirschneri in A, a clear separation between the serovars is observed. C presents the PCA for the serovars that form the class L. interrogans, which shows that the group representing L. interrogans serovar Bataviae can be completely separated, since the other clusters cannot be separated.
Identification results of 22 leptospiral field isolates by MALDI-TOF MS and 16S rRNA gene sequencing.
| Strain identification | Genome species (16S rRNA Identification) | Serogroup | MALDI-TOF-MS Identification | |
|---|---|---|---|---|
| Species | Score values | |||
| Ranarum | L. biflexa | Semaranga | L. biflexa | 2.355 |
| M85/06 | L. interrogans | L. interrogans | 2.565 | |
| M46/07 | L. interrogans | Icterohaemorrhagiae | L. interrogans | 2.070 |
| M67/07 | L. interrogans | Icterohaemorrhagiae | L. interrogans | 2.535 |
| M71/07 | L. interrogans | Icterohaemorrhagiae | L. kirschneri | 2.643 |
| M5/90 | L. interrogans | Icterohaemorrhagiae | L. interrogans | 2.342 |
| M64/06 | L. interrogans | Icterohaemorrhagiae | L. interrogans | 2.542 |
| 61H | L. kirschneri | Pomona | L. kirschneri | 1.898 |
| M110/06 | L. kirschneri | L. kirschneri | 1.866 | |
| 16CAP | L. meyeri | Grippotyphosa | L. meyeri | 2.828 |
| 19CAP | L. meyeri | Grippotyphosa | L. meyeri | 3.000 |
| LO9 | L. santarosai | L. santarosai | 2.574 | |
| M52/08-12 | L. santarosai | L. santarosai | 2.359 | |
| M52/08-19 | L. santarosai | L. santarosai | 1.833 | |
| U160 | L. santarosai | L. santarosai | 2.017 | |
| U164 | L. santarosai | Tarassovi | L. santarosai | 2.093 |
| An776 | L. santarosai | Bataviae | L. santarosai | 2.366 |
| 10ACAP | L. santarosai | Grippotyphosa | L. santarosai | 2.525 |
| 6BCAP | L. santarosai | Grippotyphosa | L. santarosai | 2.457 |
| 21CAP | L. santarosai | Grippotyphosa | L. santarosai | 2.614 |
| M4/98 | L. santarosai | Sejroe | L. santarosai | 2.370 |
| BOV 6 | L. santarosai | Sejroe | L. santarosai | 2.434 |
Collection at the Bacterial Zoonoses Laboratory, Department of Veterinary Preventive and Animal Health of School of Veterinary Medicine and Animal Science, São Paulo University, Brazil.
Principal component analysis (PCA) using tools ClinProTool™. In (A), PCA of strains analyzed, for data standardization by species, data from different serovars were used. In (B), we have PCA of different serovars of the L. kirchneri and in (C), we have PCA of different serovars of L. interrogans.
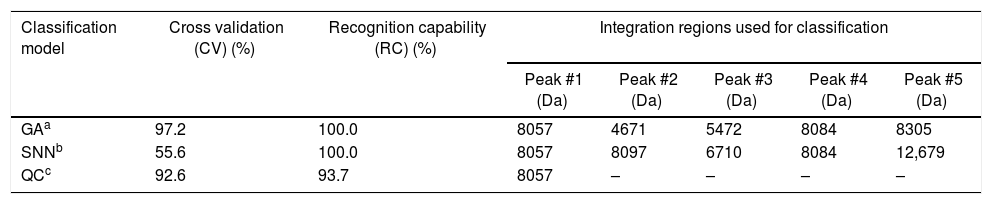
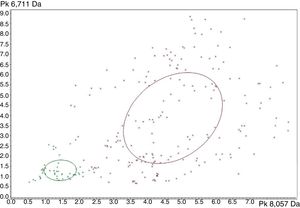
The three classification models from ClinProTools™ showed RC values ≥90% in the discrimination of L. interrogans and L. kirschneri. The best results were provided by the GA model, with RC and CV values of 100% and 97%, respectively. Details of these values are shown in Table 3. The strain distribution maps based on the GA model show that L. interrogans and L. kirschneri can be distinguished based on their peptide mass fingerprints, the best separating peaks of the current statistic sort order are displayed in Fig. 4.
Complete results derived from the classification models.
| Classification model | Cross validation (CV) (%) | Recognition capability (RC) (%) | Integration regions used for classification | ||||
|---|---|---|---|---|---|---|---|
| Peak #1 (Da) | Peak #2 (Da) | Peak #3 (Da) | Peak #4 (Da) | Peak #5 (Da) | |||
| GAa | 97.2 | 100.0 | 8057 | 4671 | 5472 | 8084 | 8305 |
| SNNb | 55.6 | 100.0 | 8057 | 8097 | 6710 | 8084 | 12,679 |
| QCc | 92.6 | 93.7 | 8057 | – | – | – | – |
Results obtained by analyzing of ClinProTools.
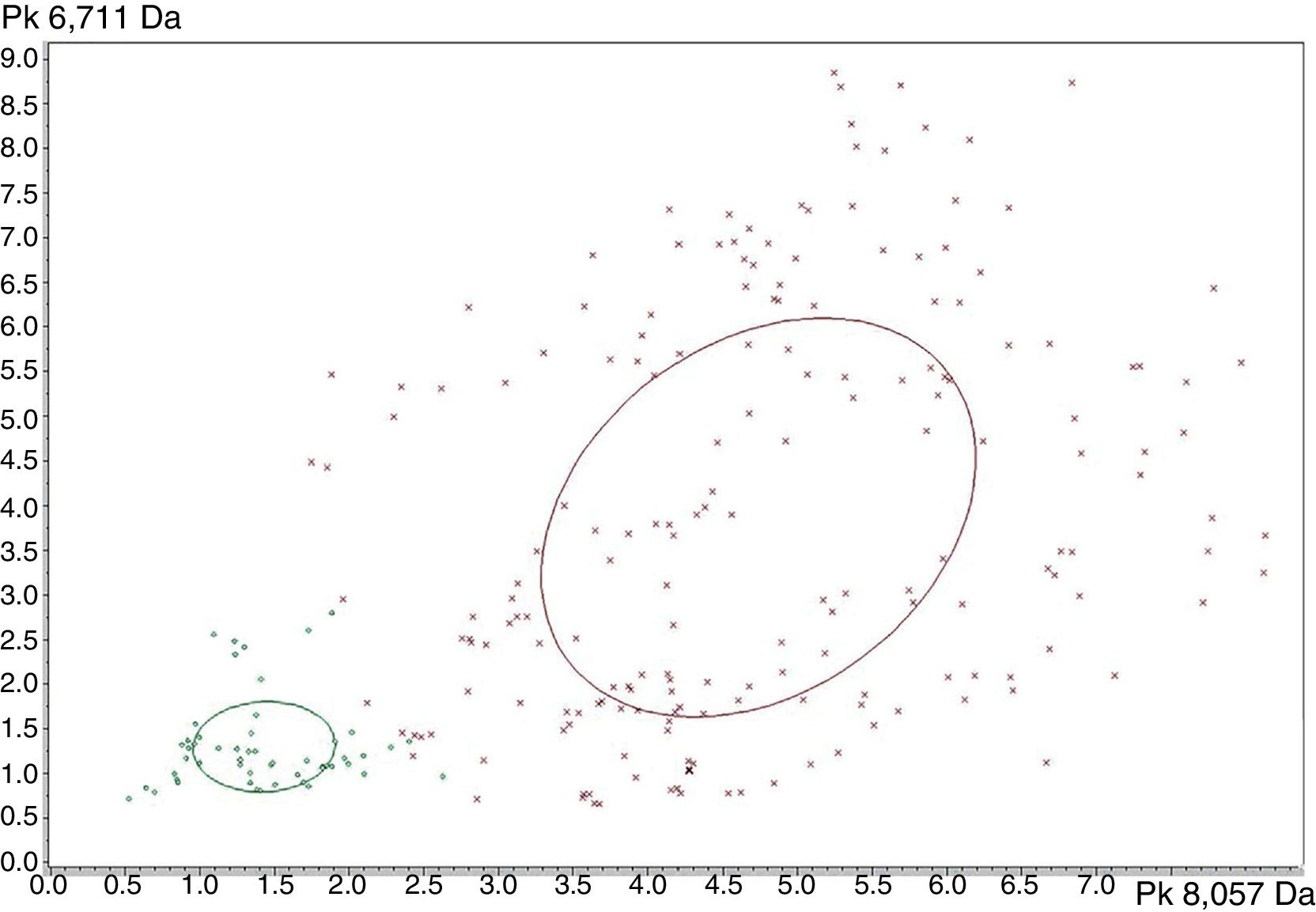
Strain distribution map corresponding to Leptospira interrogans (red) and Leptospira kirschneri (green). The x-axis shows the peak area/intensity values with respect to the most relevant peak (8057Da) to distinguish Leptospira interrogans (red) from Leptospira kirschneri (green). The y-axis shows the peak area/intensity values with respect to the peak with (6711Da) from Leptospira interrogans (red) and Leptospira kirschneri (green). The ellipses represent the spectra with greater distinction between the two groups, whereas prominent peaks in the x-axis and y-axis.
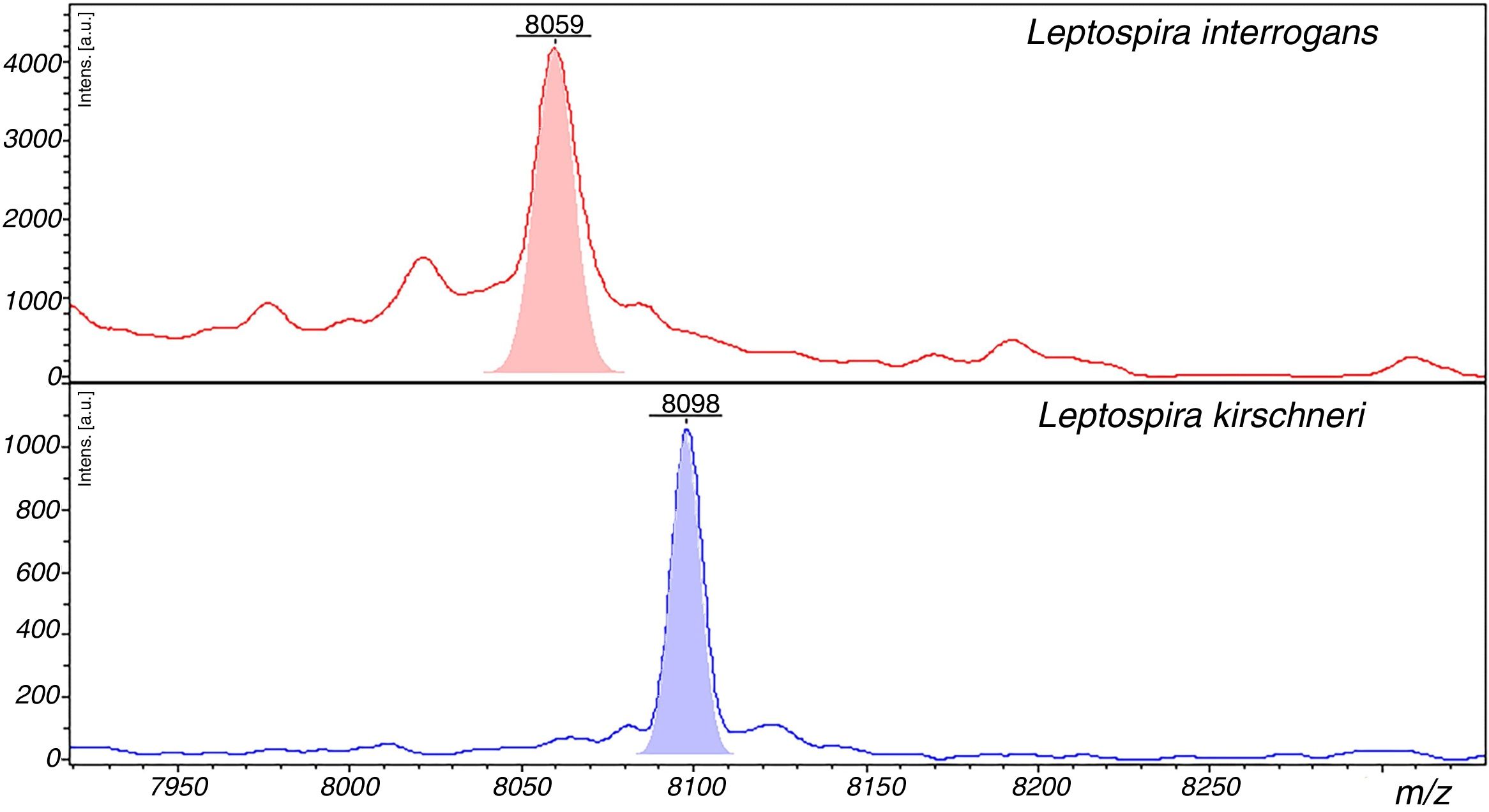
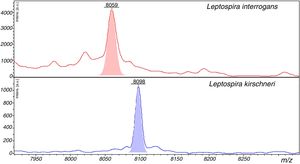
The peaks with the highest AUC (>0.9) to discriminate between L. interrogans and L. kirschneri using ClinProTools™ were 3074m/z, 3090m/z, 3118m/z, 6710m/z and 8059m/z. However, the performances of these peaks for the discrimination of the two groups using the FlexAnalysis™ software validation showed that only the peak at 8059m/z had an AUC>0.9, with sensitivity and specificity of 98.1% and 95.5%, respectively. The SN cut-off values of the peak 8057m/z peak for the discrimination of the for L. interrogans (below the cut-off) and for L. kirschneri (above the cut-off) was 7.0. The ClinProTools™ and single-peak analysis results for the differentiation of L. interrogans from L. kirschneri are summarized in Table 4 and exemplified in Fig. 5.
Single-peak analysis for the discrimination of L. kirschneri and L. interrogans.
| Peaks (m/z) | ClinProTools | FlexAnalysis | Sensitivity (%) | Specificity (%) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| AUCa | Daveb | PWKWc | PADd | Avee | Avef | AUCg | Cutoff | |||
| 8059 | 0.99 | 3.68 | 0 | 0.00226 | 16.72 | 20.63 | 0.99 | 6.96 | 98 | 95 |
| 3090 | 0.92 | 2.90 | <0.0001 | 0.000001 | 7.81 | 7.51 | 0.74 | 3.93 | 96 | 48 |
| 3074 | 0.91 | 4.14 | <0.0000 | <0.000001 | 7.13 | 8.04 | 0.83 | 5.41 | 100 | 59 |
| 3118 | 0.90 | 12.03 | <0.0001 | <0.000001 | 8.38 | 9.47 | 0.83 | 14.00 | 100 | 56 |
| 6710 | 0.87 | 2.85 | 0 | <0.000001 | 14.26 | 16.36 | 0.87 | 3.22 | 85 | 86 |
Peaks with the best performances according to ClinProTools™ and FlexAnalysis™ softwares.
Representative spectra of Leptospira interrogans and Leptospira kirschineri. The representative peaks that allow differentiation of the strains in the spectra are showed, in red for L. interrogans, and in blue for L. kirschneri. The peak with m/z=8059 in L. interrogans we detected as shown in Table 3 by ClinPro Tools analysis. The peak m/z=8098 in L. kirschineri was previously identified by Rettinger et al.10
During leptospirosis outbreaks, Leptospira species identification is an essential step for tracking and controlling the pathogen transmission. The determination of a serovar may be insufficient as different species may have the same serovar but may be distinct in their ability to cause mammalian infection.13 DNA sequencing is an alternative method for the identification of Leptospira species, although it can be a costly, time-consuming and labor-intensive technique. MALDI-TOF MS has been successfully applied in the identification of Leptospira species, yielding comparable results to 16S rRNA sequencing, with fast, reproducible and less costly protocols.6–8 However, the creation of an in-house MSP database with well-identified strains remains necessary until an updated commercial database with Leptospira MSPs becomes available. A score-oriented dendrogram produced by Biotyper™ software with 31 MSPs clustered the strains according to their pathogenicity clearly separated pathogenic and non-pathogenic Leptospira strains into different nodes. Our analysis reproduced the results that have been reported by other centers that also constructed in house Leptospira MSP databases. Moreover, all field isolates had the correct species assigned, with scores above 2.0, which ensures the quality of our MSP database for Leptospira species ID. The distinction of L. interrogans and L. kirschneri using ClinProTools™ and single-peak analysis is also noteworthy. Although MALDI-TOF MS has already been successfully applied in Leptospira genus and species identification,6–8 the misidentification of closely related species, such as L. interrogans and L. kirschneri, has also been reported and represents an important challenge in the implementation of MALDI-TOF MS for Leptospira identification. Here we observed that with proper analysis, Leptospira species can be distinguished based on their peptide mass fingerprints.
The ClinProTools™ software is a biomarker analyzer that has been widely applied in microbiology, providing a rapid and cost-saving method for epidemiological clustering, strain typing and subspecies identification.14–16 Using both ClinProTools™ and single-peak analysis with FlexAnalysis™ has provided higher discriminatory power to detect bio-marker peaks.14–17 Our results corroborate previous findings that one isolate biomarker with 8000–8100Da can effectively distinguish the closely related pathogenic species L. interrogans from L. kirschneri.10 Indeed, we further described the SN cut-off value that has to be adopted to accurately differentiate these two taxa by a simple inspection of the mass spectrum. Recently, L. kirschneri serovar Mosdok was, for the first time, linked to human leptospirosis in the southern hemisphere; therefore, rapid species ID using MALDI-TOF MS may be the first step to implement control strategies.
Conflicts of interestThe authors declare no conflicts of interest, even during the item proofs.
This work was supported by Fundacao de Amparo a Pesquisado Estado de São Paulo (FAPESP—Projects – 12/50191-4R) and Conselho Nacional de Desenvolvimento Cientifico e Tecnologico (CNPq—Projects – 443978-2014-0 and 467478-2014-7). L.Z.M. is recipient of a PhD fellowship from FAPESP (2013/17136-2).