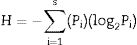Arbuscular mycorrhizae (AM) fungi play a crucial role in the growth of soybean; however, the planting system employed is thought to have an effect on AM fungal communities in the rhizosphere. This study was performed to explore the influence of continuous soybean cropping on the diversity of Arbuscular mycorrhizal (AM) fungi, and to identify the dominant AM fungus during the seedling stage. Three soybean cultivars were planted under two and three years continuous cropping, respectively. The diversity of AM fungi in the rhizosphere soil at the seedling stage was subsequently analyzed using polymerase chain reaction (PCR)-denaturing gradient gel electrophoresis (DGGE). The results showed that an increase in cropping years improved the colonization rate of AM in all three soybean cultivars. Moreover, the dominant species were found to be Funneliformis mosseae and Glomus species. The results of cluster analysis further confirmed that the number of years of continuous cropping significantly affected the composition of rhizospheric AM fungal communities in different soybean cultivars.
Soybean is the fourth most important grain cultivar in the world, and an important oil crop in China. Soybean contains active substances that are beneficial to human health such as soy proteins, isoflavones, saponins and oligosaccharides.1 Considerable attention has recently been directed toward the increasingly severe problem of root rot in soybean under intensified cultivation. Continuous cropping has resulted in an increase in diseases and pests, poor soil chemical and physical properties, a drastic reduction in yield and, in some cases, total crop failure.2–4 Tackling the yield reductions resulting from continuous cropping has therefore become a key issue in the soybean industry. Recent studies suggest that continuous cropping of soybean leads to drastic changes in the rhizosphere microflora, with a gradual transition from bacterial-dominant highly fertile soil to fungal-dominant low fertile soil, and therefore, microbiota dominated by fungi.5 Moreover, the concentration of root exudates such as phenolic acids is significantly correlated with fungal quantity, while increased levels of soil organic compounds (sugars, amino acids and organic acids) as a result of continuous cropping play a facilitative role in the growth of root rot pathogens. Since soil sterilization treatments have no effect on fungal spores, a gradual transition to a dominant fungal community is subsequently observed.6
Arbuscular mycorrhizae (AM) fungi are the most important type of endotrophic mycorrhiza, and are associated with the majority of terrestrial plants. AM fungi are a group of ubiquitous microbes in nature, and are one of the major types in the rhizosphere,7–10 significantly improving the absorption and utilization of nutrients by host plants. AM fungus also stimulate growth, increase stress and disease resistance, and promote community succession and ecosystem stability.11 AM fungi are known for several effects and can be used as bio-fertilizer.12–16 Application of AM fungi is widely recognized as important for many crops including soybean.17,18 AM fungi applied to soybean can improve the absorption of nutrients in the host plants, improve the nitrogen fixation capability in Rhizobia and also the colony structure of the rhizospheric econiche, thus increasing the yield and economic benefits of soybean.19,20 The antagonistic effects of AM fungus on the problems associated with continuous cropping have therefore been examined.21,22 However, the diversity of AM fungi in rhizospheric soil with different soybean cultivars under different continuous cropping regimes remains largely unknown.
In this study, three typical soybean cultivars (Heinong 37, Heinong 44 and Heinong 48) grown widely in Heilongjiang province, China, were selected to examine the diversity of AM fungi in the rhizosphere during continuous cropping. Polymerase chain reaction-denaturing gradient gel electrophoresis (PCR-DGGE) was employed with the aim of identifying dominant AM Fungi communities. The findings provide a theoretical basis for further studies of the relationship between AM fungi and soil-borne pathogens.
Materials and methodsExperimental plotsThis study was carried out at the Experimental Station of the Research Institute of Sugar Industry, Harbin Institute of Technology, China (125°42′–130°10′E, 44°04′–46°40′N). Soybean cultivars were planted in the same plot and subjected to two or three years continuous cropping, respectively. Management techniques in the trial field were the same as those in standard production fields. Organic matter, potassium, phosphorus and nitrogen contents of the soil were determined using the Ignition method, NaOH melt-flame photometry method, alkali fusion-Mo-Sb Anti spectrophotometric method and Kjeldahl method, respectively.
Soybean cultivarsThree typical soybean cultivars planted widely in Heilongjiang Province were used: ‘Heinong 44’ (high oil, 36% average protein content, 23% average fat content, designated HN44), ‘Heinong 37’ (intermediate cultivar, 40% average protein content, 20% average fat content, designated HN37), and ‘Heinong 48’ (high protein, 45% average protein content, 19% average fat content, designated HN48). Each cultivar was subjected to two and three years continuous cropping, respectively, as follows: 2 years continuous cropping of HN37 (designated C2HN37), 2 years continuous cropping of HN44 (C2HN44), 2 years continuous cropping of HN48 (C2HN48), 3 years continuous cropping of HN37 (C3HN37), 3 years continuous cropping of HN44 (C3HN44) and 3 years continuous cropping of HN48 (C3HN48). The protein content of the soybean seeds was determined using the Kjeldahl method and the fat content determined using the Soxhlet extraction method.
Harvesting and processing of samplesSoybean samples were collected after 30 d (at the seedling stage) along with soil samples. Three soybean plants per cultivar were randomly selected from each treatment by digging to a depth of 10–20cm. The plants were shaken gently and any soil adhering to the root surface 1–3mm removed using a brush as rhizosphere soil. The three soil samples per treatment were subsequently mixed as a collective sample. Samples were then air dried then stored at 4°C until use. The roots were then washed for soil sample collection, and three randomly selected segments removed with scissors and used as a collective sample. All treatments were sampled in triplicate. Roots were then washed in distilled water and placed in FAA fixative and the level of AM fungal colonization determined microscopically. Roots were stained using alkali hydrolyzable and acid fuchsin, then stored at 4°C until use. Root and soil samples from each cropping system were denoted RL2 and RL3, and SL2 and SL3 to represent 2 and 3 years continuous cropping, respectively.
Determination of the rate of AM fungal colonizationFifty fibrous root segments from each root sample were selected randomly, stained, prepared on slides and examined microscopically to observe the level of AM colonization. Colonization rates of each fibrous root segment were assigned values of 0, 10, 20, 30, 40 and 100% based on the microscopic observations and the colonization frequency (F) of each cultivar under each cropping system calculated as follows:
AM colonization intensity (M) was then calculated statistically for each fibrous root segment as follows23:
Mean colonization rates and standard deviations were calculated using Excel software to give a variation rule of colonization frequency and intensity.
PCR-DGGETotal DNA in the soybean roots and rhizosphere soil was extracted using the cetyl trimethyl ammonium bromide (CTAB) method and the Omega genomic DNA extraction series E.Z.N.A.® Soil DNA kit. The nested-PCR procedure was performed as described by Jie.24
In the first amplification, primers GeoA2 (5′-CCAGTAGTCATATGCTTGTCTC-3′) and Geo11 (5′-ACCTTGTTACGACTTTTACTTCC-3′) were used to amplify the fungal 18S rDNA sequence as follows: initial denaturation at 94°C (4min) followed by 30 cycles of denaturation at 94°C (1min), annealing at 54°C (1min), and extension at 72°C (2min), ending with final extension at 72°C (7min).
Products from the first amplification were subsequently diluted 1/100 and used as template for a second amplification using the AM fungal specific primer AM1 (5′-GTTTCCCGTAAGGCGCCGAA-3′) combined with the universal eukaryotic primer NS31-GC (5′-TTGGAGGGCAAGTCTGGTGCC-3′). Amplification was performed in a thermal cycler as follows: initial denaturation at 94°C (2min) followed by 30 cycles of denaturation at 94°C (45s), annealing at 65°C (1min) and extension at 72°C (45s), ending with final extension at 72°C (7min). All PCR amplification reactions were conducted in a 20μL volume containing 2.0μL 10× PCR buffer (with Mg2+), 2.0μL 2.5mM dNTPs, 0.5μL primer 1 (10μM), 0.5μL primer 2 (10μM), 0.1μL Taq DNA polymerase, and 2μL DNA template. Amplification products were separated by gel electrophoresis on 1.0% (W/V) agarose gel in TAE buffer.
Products from the second PCR amplification were again diluted 1/100 and used as template for a third reaction using the primers NS31-GC (5′-TTGGAGGGCAAGTCTGGTGCC-3′) and Glol (5′-GCCTGCTTTAAACACTCTA-3′). Amplification was performed in a thermal cycler as follows: initial denaturation at 94°C (2min) followed by 30 cycles of denaturation at 94°C (45s), annealing at 55°C (1min) and extension at 72°C (45s), ending with final extension at 72°C (7min). Amplification products were separated by gel electrophoresis on 1.2% (w/v) agarose gel in TAE buffer.
A total of 3.0μL of the third PCR products of root and soil samples from each soybean cultivar was used for DGGE analysis. DGGE conditions were optimized by repetitive trials on 8% polyacrylamide gel containing denaturing agents. DGGE conditions were as follows: denaturant range, 40–65%; electrophoresis temperature, 60°C; pre-running voltage, 50V; electrophoresis voltage, 130V; electrophoresis time, 9h; and silver staining, 15min.
Phylogenetic analysisThe PCR products were cloned and transformed into a vector, and positive clones selected and sequenced. The genetic relationships between the sequences were subsequently determined, and the sequences compared to known sequences of similar strains in GenBank. MEGA 5.1 was used to construct a phylogenetic tree based on the neighbor-joining method. Bootstrap resampling analysis of 1000 replicates was performed to determine the confidence of the tree topologies.
DGGE and cluster analysisDiversity is often indicated by sample richness, dominance, and the diversity index. The richness of the DGGE patterns was compared using a Gel-Pro Analyzer, and diversity calculated using the equation below. Cluster analysis of the DGGE bands was subsequently performed using Quantity One 4.62 software.
Richness: number of bands per lane in the DGGE patterns.
Diversity index (H value): comprehensively reflected by species richness and species evenness, indicated by the Shannon-Weiner index (large H values indicating higher diversity):
where S is the number of bands in the sample DGGE fingerprint pattern and Pi the dominance of the ith band.25Statistical analysisAll values are expressed as the mean±SD. Analysis of variance (ANOVA) was used to determine the significance of differences between AMF communities of the three soybean cultivars using SPSS 19.0. Values of p<0.05 were considered statistically significant.
ResultsPhysicochemical parameters of the analyzed soilMajor physical and chemical soil indicators in the plots included: organic matter, 26.13gkg−1; total N, 1.69gkg−1; total K, 25.4gkg−1; total P, 5.5gkg−1; alkali-hydrolyzable N, 133.1mgkg−1; rapidly available P, 13.14mgkg−1, rapidly available K, 206mgkg−1; and pH, 7.0.
Effect of years of continuous cropping on AM fungal colonization of each soybean cultivarAM fungal colonization rates of the soybean seedlings in each plot under each continuous cropping system were calculated statistically (Table 1). Colonization rates of each cultivar were higher under three years continuous cropping than two years, suggesting that an increase in cropping years is beneficial for AM fungi colonization of soybean roots. This was perhaps related to transition of the soil community structure from bacterial to fungal during continuous cropping. The AM fungal population will subsequently increase with an increasing proportion of fungi in the rhizosphere community, thus leading to a natural increase in AM fungal colonization rates.
Arbuscular mycorrhizal (AM) fungal colonization (%) of seedlings of three soybean cultivars under continuous cropping.
| Years of continuous cropping | HN37 | Cultivar HN44 | HN48 |
|---|---|---|---|
| Two | 28.9±2.5a | 19.9±1.3a | 17.8±1.2a |
| Three | 39.5±2.8b | 33.1±2.2b | 31.2±2.5b |
Note: Lowercase letters in the same column indicate a significant difference at p<0.05.
Colonization rates at the seedling stage differed significantly between cultivars under the same cropping system (Table 2), with Heilong 37 showing the highest colonization rate. Nutrient use differs between soybean cultivars, and continuous cropping amplifies the differences in rhizosphere nutrients used. Rhizosphere microflora populations therefore also differ with different soybean cultivars, leading to interspecies differences in AM fungal colonization rates.
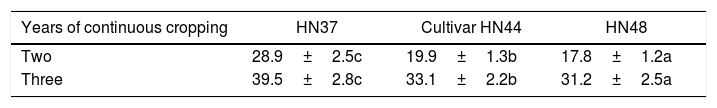
Variance analysis of the effects of two and three years continuous cropping on AM fungal colonization (%) of three soybean cultivars.
| Years of continuous cropping | HN37 | Cultivar HN44 | HN48 |
|---|---|---|---|
| Two | 28.9±2.5c | 19.9±1.3b | 17.8±1.2a |
| Three | 39.5±2.8c | 33.1±2.2b | 31.2±2.5a |
Note: Lowercase letters in the same line indicate a significant difference at p<0.05.
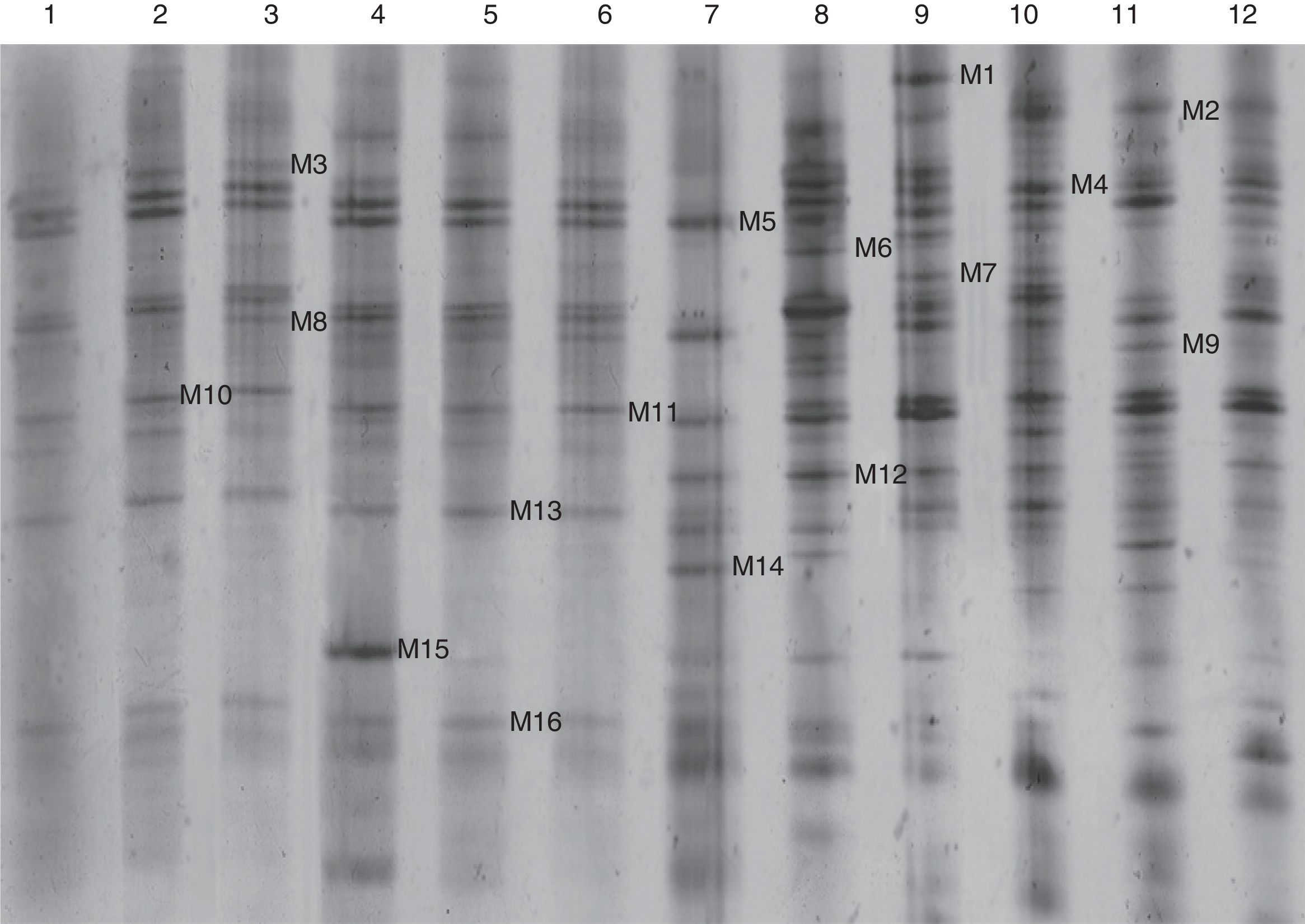
PCR products from the NS31/Glol region of root and soil samples of the three soybean cultivars were subjected to DGGE analysis. DGGE and silver staining revealed diverse banding patterns (Fig. 1), which were clearly separated by different fingerprint patterns and different degrees of brightness. Sixteen bands, M1 through M16 (M represents microbial), that occurred at a high frequency and were clearly separated were selected for further isolation. The recovered bands were re-amplified and reanalyzed by DGGE. Amplified PCR products were then ligated to the pGEM-T vector (Tiangen, GER) and transformed into Escherichia coli DH 5 (AS, CHN) competent cells. Positive clones were screened, re-amplified and analyzed once more using DGGE to verify that the DNA segment inserted into the plasmid represented single-band DNA. Positive clones complying with the DGGE detection requirements were selected for sequencing.
Denaturing gradient gel electrophoresis (DGGE) banding patterns of root and soil samples from three soybean cultivars after two and three years continuous cropping. Lanes 1–6: root samples; lanes 7–12: soil samples.
1: HN37, two years cropping; 2: HN37, three years cropping; 3: HN44, two years cropping; 4: HN44, three years cropping; 5: HN48, two years cropping; 6: HN48, three years cropping; 7: HN37, two years cropping; 8: HN37, three years cropping; 9: HN44, two years cropping; 10: HN44, three years cropping; 11: HN48, two years cropping; 12: HN48, three years cropping.
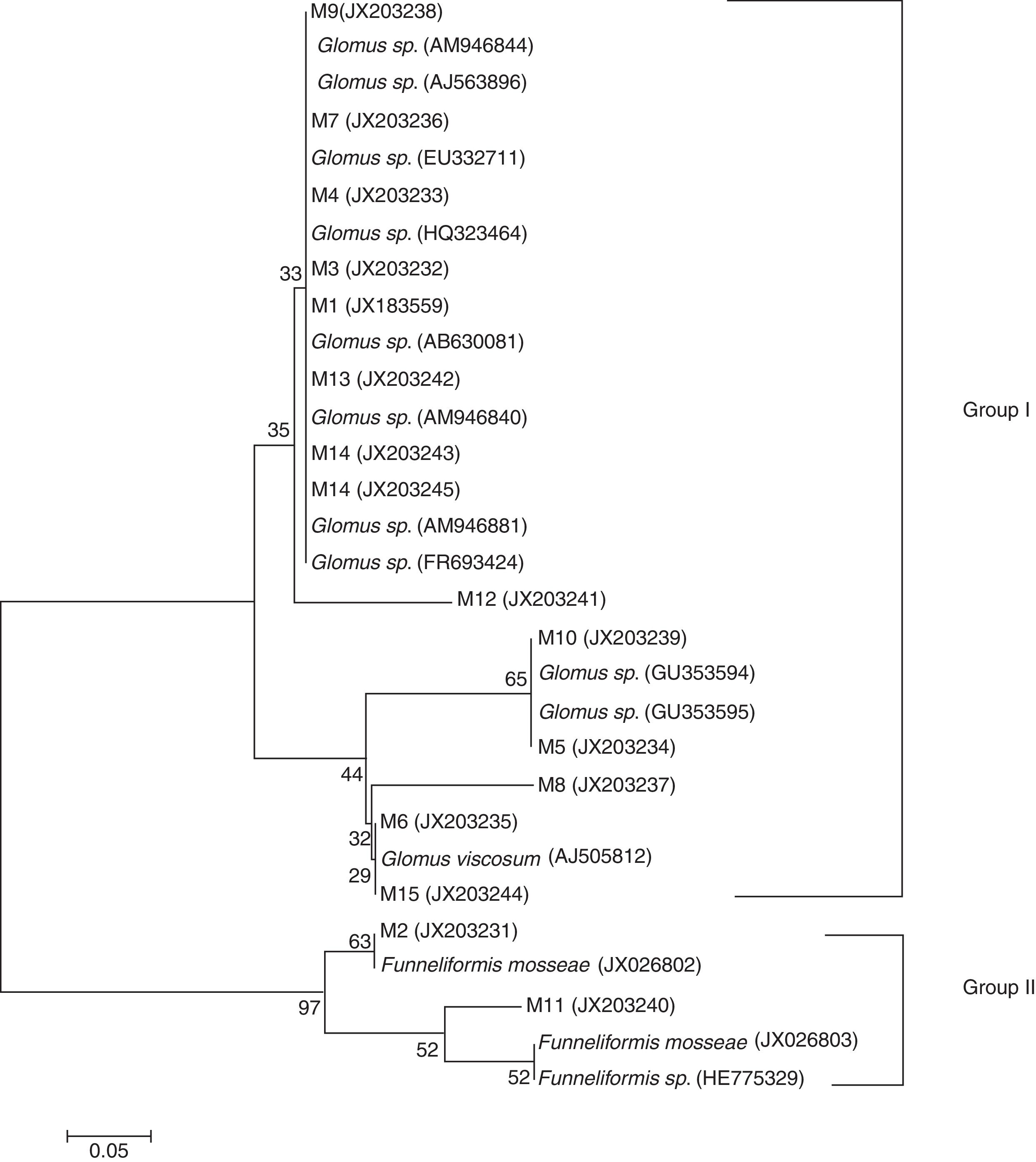
Sequencing revealed that the 16 DNA fragments were 234bp in length. They were subsequently compared with known strains in the GenBank sequence database. The results revealed >97% similarity, preliminarily suggesting the presence of a dominant strain. Microflora represented by the 16 bands all belonged to AM (Fig. 2). M2 and M11 fragments belonged to Funneliformis mosseae with similarities of >98%, while those represented by the remaining 14 bands belonged to species in the genus Glomus. M6 and M15 belonged to Glomus viscosum with similarity of up to 99%. Bands representing these dominant AM fungus were statistically assigned to 16 strains. Based on these sequencing results, the dominant AM fungi in the rhizosphere communities were deemed to be Glomus and Funneliformis mosseae. MEGA 5.1 was subsequently used to construct a phylogenetic tree (Fig. 2).
Fig. 2 shows that the 16 bands generated by DGGE are mainly representative of two types of microflora: Glomus spp. and Funneliformis mosseae. The two types are dominant AM fungi. The first group belongs to the genus Glomus, and includes Glomus viscosum; the second group comprises the species Funneliformis mosseae.
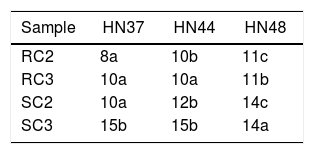
Effect of continuous cropping on the population structure of AM fungus communities in the rhizosphere of three soybean cultivarsStatistical analysis of richness data of both the root and soil samples revealed that under three year cropping all three cultivars showed higher richness of AM fungi compared to two years cropping. These results suggest that increased years of continuous cropping is beneficial for improving the richness of AM fungal communities. Analysis of variance further revealed that the relative richness of AM fungal communities in soybean cultivars at the seedling stage for a given cropping year were significantly different (Table 3).
Richness of arbuscular mycorrhizal fungi in the rhizosphere of three soybean cultivars on different cropping years.
| Sample | HN37 | HN44 | HN48 |
|---|---|---|---|
| RC2 | 8a | 10b | 11c |
| RC3 | 10a | 10a | 11b |
| SC2 | 10a | 12b | 14c |
| SC3 | 15b | 15b | 14a |
RC2: root sample, 2 years continuous cropping; RC3: root sample, 3 years continuous cropping; SC2: soil sample, 2 years continuous cropping; SC3: soil sample, 3 years continuous cropping.
HN37: Heinong 37; HN44: Heinong 44; HN48: Heinong 48.
Note: Lowercase letters in the same row indicate a significant difference at p<0.05.
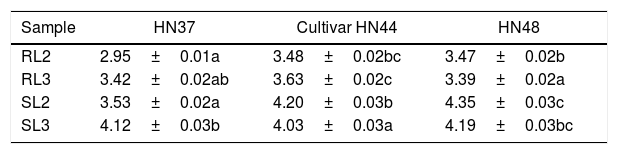
Under three years cropping, root and soil samples of all three cultivars had a higher AM fungal diversity index compared to two years cropping based on statistical diversity index data. These findings suggest that continuous cropping is beneficial for improving the diversity index of AM fungal communities. Analysis of variance further revealed that the diversity indexes of AM fungi of each soybean cultivar for the same cropping years were significantly different (Table 4). For example, RL3 of HN37 did not significantly differ from RL3 of HN48, but both were significantly different from HN44. This finding suggests that the AM fungal communities in HN48 roots had a higher diversity index, and therefore, a more complex AM population structure.
Analysis of variance of the effects of two and three years continuous cropping on arbuscular mycorrhizae (AM) fungal community diversity in three soybean cultivars.
| Sample | HN37 | Cultivar HN44 | HN48 |
|---|---|---|---|
| RL2 | 2.95±0.01a | 3.48±0.02bc | 3.47±0.02b |
| RL3 | 3.42±0.02ab | 3.63±0.02c | 3.39±0.02a |
| SL2 | 3.53±0.02a | 4.20±0.03b | 4.35±0.03c |
| SL3 | 4.12±0.03b | 4.03±0.03a | 4.19±0.03bc |
Note: Lowercase letters in the same row indicate a significant difference at p<0.05.
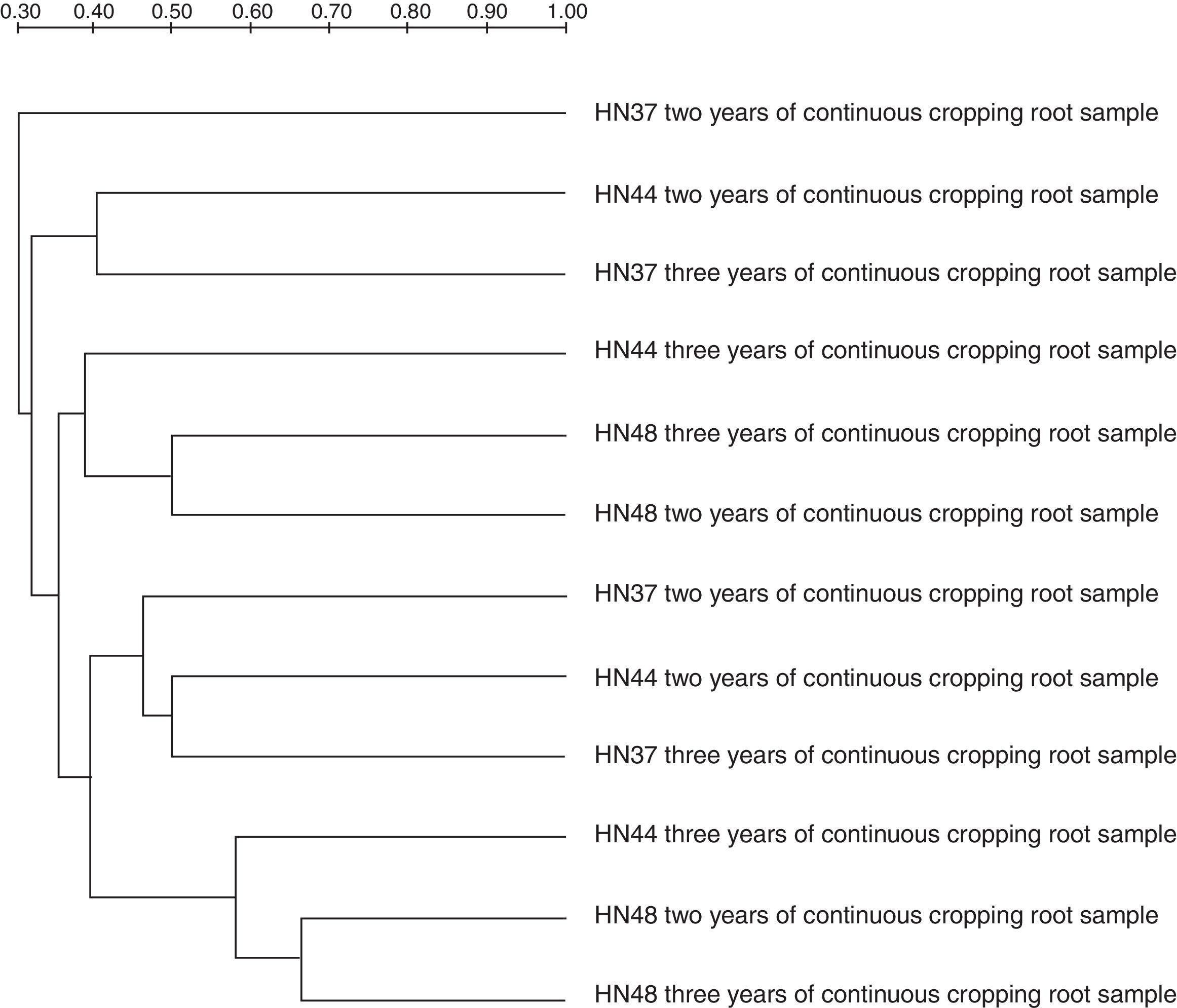
Cluster analysis of the DGGE profiles showed that the AM fungal communities were clustered into eight groups (Fig. 3). Group I included the root samples of HN37 under two years cropping); Group II the root samples of HN37 under three years cropping and HN44 under two years cropping; Group III the soil samples of HN44 under three years cropping; Group IV the soil samples of HN48 under two and three years cropping; Group V the soil samples of HN37 under two years cropping; Group VI the soil samples of HN37 under three years cropping and HN44 under two years cropping; Group VII the root samples of HN44 under three years cropping; and Group VIII the root samples of HN48 under two and three years cropping. The findings suggest that AM fungal communities were affected by the soybean cultivar, cropping duration and micro-habitat (soil or root). That is, these communities were cultivar-specific and dependent on the cropping duration.
DiscussionIn this study, morphological analysis and molecular genetics were combined to investigate the effect of continuous cropping on AM fungal colonization and population structure in roots and rhizosphere soil of three soybean cultivars. The dominant species were found to be Funneliformis mosseae and Glomus species. A significant effect of years of continuous cropping on AM fungi of the different soybean cultivars was also revealed, supporting further studies of the relationships between AM fungi and soil-borne disease pathogens resulting from continuous cropping.21,22
The number of continuous cropping years affected the AM fungal population structure of both the roots and rhizosphere, while DGGE profiles showed that the population structure was more complex under three years cropping compared to two. These findings suggest that continuous cropping is beneficial to AM fungal diversity. However, DGGE patterns of fungal richness differed at different stages of soybean growth. At the branching stage, bands M10, M11, M12, M14 and M15 were observed in samples from both two and three years cropping. This suggests that the microflora represented by these five bands was relatively stable in the population structure of the rhizosphere soil.26 However, at the seedling stage, bands M2, M5, M7 and M11 were observed. It fully illustrated that the two studies could be complimentary to each other. Results at the podding stage, the third most important stage of soybean growth, now need to be confirmed.
Under different cropping years, the AM fungi also showed different colonization rates, with rates under three years cropping higher than those under two years cropping. This finding suggests that continuous cropping is beneficial to AM growth and colonization. A number of studies suggest that AM fungi form mycorrhiza more efficiently in barren soil.27–29 Under short-term continuous cropping (two to three years), the amount and diversity of AM fungi in the soil increased, suggesting the ability to ease the deleterious effects of continuous cropping on soybean growth. Population structures of the AM fungal communities in the rhizosphere soil significantly differed between two and three years continuous cropping, and these different AM fungi were not completely consistent with the proportion of AM fungal communities in the soil. This suggests that different soybean cultivars have different AM fungi colonization efficiencies. Understanding the variation in AM microflora in rhizosphere soil with different soybean cultivars is therefore important since these communities serve as a microbial fertilizer, minimizing the negative effects of continuous cropping and promoting growth.
This study revealed that the total richness of AM fungi in root and soil samples of the three soybean cultivars (HN37, HN44 and HN48) was higher under three years cropping than two. This suggests that continuous cropping causes a change in soil nutrients, soil microorganisms and soil enzyme.30–32 For example, continuous cropping may cause a significant decrease in pH, causing a change in soil from neutral to acidic, thereby favoring the fungal growth and inhibiting the breeding of bacteria and actinomycete. Overall, this leads to fungal dominance in the soil.
Despite our findings, the results of nested-PCR revealed that the length of the PCR products gradually decreased Flaws in the primers used possibly caused the amplification of non-AM fungi, even though many studies suggest the feasibility of the selected primers.30,31 Other authors have reported that the AM fungi primer failed to amplify AM fungi belonging to Archaeosporaceae and Paraglomaceae.32 Our sequencing results suggest that the AM fungal communities were dominated by bands belonging to Glomus species, with no bands containing species belonging to the above two families. Glomus was therefore considered the dominant genus of AM fungi in the soybean rhizosphere, consistent with previous studies.33,34
Conflicts of interestThe authors declare no conflicts of interest.
This work was supported by grants from the National Natural Science Foundation of China (No. 31570487) and the Key Project of Horizontal Subject of Heilongjiang East University (No. HDFHX160103).