Fungi is a well-known model used to study drug metabolism and its production in in vitro condition. We aim to screen the most efficient strain of Cunninghamella sp. among C. elegans, C. echinulata and C. blakesleeana for bromhexine metabolites production. We characterized the metabolites produced using various analytical tools and compared them with mammalian metabolites in Rat liver microsomes (RLM). The metabolites were collected by two-stage fermentation of bromhexine with different strains of Cunninghamella sp. followed by extraction. Analysis was done by thin layer chromatography, high performance thin layer chromatography, Fourier transform infrared spectroscopy, high performance liquid chromatography and Liquid chromatography–mass spectrometry. The role of Cytochrome P3A4 (CYP3A4) enzymes in bromhexine metabolism was studied. Fungal incubates were spiked with reference standard – clarithromycin to confirm the role of CYP3A4 enzyme in bromhexine metabolism. Three metabolites appeared at 4.7, 5.5 and 6.4min retention time in HPLC. Metabolites produced by C. elegans and RLM were concluded to be similar based on their retention time, peak area and peak response of 30.05%, 21.06%, 1.34%, and 47.66% of three metabolites and bromhexine in HPLC. The role of CYP3A4 enzyme in metabolism of bromhexine and the presence of these enzymes in Cunninghamella species was confirmed due to absence of peaks at 4.7, 5.4 and 6.7min when RLM were incubated with a CYP3A4 enzyme inhibitor – clarithromycin.
The approval and usage of drugs for human consumption need extensive pharmacokinetic and pharmacodynamics studies to determine its safety and efficacy. Drug metabolism plays an important role in the development of new drug entities to be further evaluated for pharmacological/toxicological activities. The formation of metabolite and its role in the body before excretion is important to understand drug's safety and toxicity profile. Cytochrome P450 enzyme plays an important role in the metabolism of the drugs. To signify CYP450 and its isoforms using in vitro methods the pharmacokinetic profile of potential drugs in human beings has rapidly progressed.1 Conventionally drug metabolism studies are performed on tissue culture, in vitro enzyme systems, small animal models, perfused organs, in vitro cell cultures and liver microsomal preparations. The quality and yield of the microsomes in the preparations is judged by the NADPH-dependent N- and p-hydroxylation of N-ethylaniline, protein content, and cyanide-sensitive respiration.2 Synthesis of metabolites in laboratories is tedious process therefore microorganisms can be used as a resourceful alternative to produce these metabolites. Cytochrome P450s represent the most important class of enzymes involved in phase I metabolism, being involved in 75–80% of metabolism of marketed drugs. Phase I reactions involve hydroxylation, epimerization, oxidation and reduction.3 These enzymatically catalyzed reactions alter aromatic amino functional groups. This increases the polarity and/or makes aromatic amino acid structures more accessible to phase II metabolizing enzymes. The primary superfamily of phase I enzymes are Cytochrome P450, which hold catalytic versatility.1 A relatively novel perspective in modern drug development is the use of pharmacologically active metabolites as potential resources for drug discovery and development. There are several advantages for screening drug candidates for active metabolites during drug discovery.4 By creating a so-called prodrug it is possible for the drug to reach the target area in the body and gain the pharmacological effect when it undergoes a biotransformation process, an example is drug amitriptyline. Amitriptyline is demethylated in the liver to nortriptyline which is an active metabolite to give its pharmacological action.5 Drug toxicity can arise from the biotransformation when a metabolite is electrophilic and react with e.g., a protein. A classic example is paracetamol that can cause liver damage. Damage to the liver is due to a toxic metabolite (N-acetyl-p-benzoquinone imine (NAPQI)) produced by cytochrome P450 enzymes in the liver. The drug could also cause rare and possibly fatal, skin reactions; asthma and hearing loss. This damage is averted by conjugation of paracetamol’ s reactive metabolite with glutathione but when the reserves of glutathione run low, serious liver damage can be the result.6
Microbial metabolism model evolved as one of the in vitro model to subjugate the flaws and disadvantages of other mammalian models. Microbial models can be used as a supplementary tool to imitate mammalian metabolism by minimizing the usage of animals and human subjects in the drug discovery and development process. Microorganisms such as bacteria, yeast and fungi can be used as in vitro models for successful prediction of mammalian drug metabolism with significant applications. They have shown their possible use in vitro models of drug metabolism. There are studies that have shown that fungi Cunninghamella species produce mammalian metabolites of different drugs such as meloxicam, naproxen, amitriptyline, omeprazole, clemastine and more.7–10 These fungi produce metabolizing enzymes that can perform both phase I and phase II biotransformation reactions and their metabolism of numerous substances have been investigated. C. elegans has been proven to carry at least one gene coding for a CYP enzyme closely related to the CYP51 family.11Cunninghamella species are mainly soil fungi of the Mediterranean and subtropical zones; they are rarely isolated in less temperate regions. The experimental procedure of cultivating the fungi and using them in biotransformation is simple, requires a low input of work and has a low cost.12 Different approaches can be identified for the development of microbial models for a given drug; the prediction and confirmation of metabolite production in mammals can be facilitated by prior generation of analytical standards of metabolites using the microbial model or a parallel microbial and mammalian metabolism may be conducted. In any approach, the preparative scale production of metabolites would facilitate their biological and toxicological evaluation, stereochemistry, and mechanism of formation.
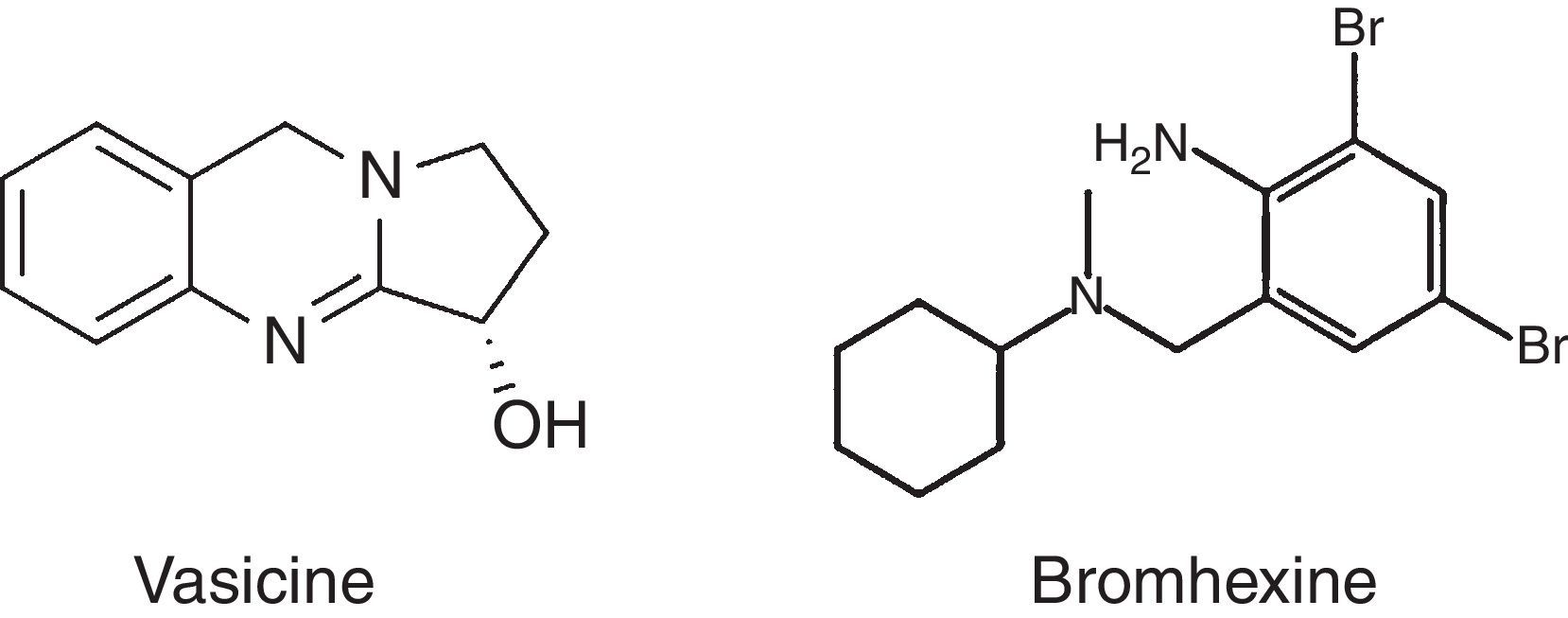
Bromhexine (2-amino-3, 5-dibromo-N-cyclohexyl-N-methylbenzylamine) is a synthetic derivative of vasicine, one of the active ingredients of the Asian plant Adhatoda vasica (Fig. 1). Bromhexine has proven its effectiveness in normalizing mucus in the respiratory tract so that a natural cough response can clear the airway. Introduced for the first time in 1963, as a secretolytic or mucolytic medicine by Boehringer Ingelheim, bromhexine became one of the most popular cough remedies. Today, it is still widely used as an over-the-counter drug. CYP3A4 enzymes are responsible for the drug's metabolism and in clinical use bromhexine is administered orally three times a day at dosage of 8 or 16mg per dose.13 Ambroxol is an active N-desmethyl metabolite of bromhexine. It is formed after deletion of a methyl group and introduction of a hydroxyl group in a para-trans position of cyclohexyl ring. It gains several important pharmacological properties like surfactant stimulatory, anti-inflammatory, anti-oxidant, and local anesthetic effect besides the muco-kinetic and muco-ciliary effects of the parent compound bromhexine.12 Some drugs that inhibit CYP3A4 activity increases the plasma concentrations of the CYP3A4 substrate drug. Drugs, such as clarithromycin, itraconazole, and ketoconazole, are potent inhibitors of CYP3A4; these drugs may have markedly reduced CYP3A4 activity in patients.14 The metabolism of bromhexine has been studied using pig hepatocyte cultures. They maintain both phase I and phase II biotransformation reactions and considered to be an established in vitro model. Phase I reactions, i.e. hydroxylation and demethylation occurs fast whereas hydroxylated/demethylated and aminal hydroxylated metabolites formation from multiple-step reactions takes longer time. Besides metabolites formed in vivo, three unknown components were also detected.15
The aim of the present work was to evaluate the formation of metabolites of bromhexine using three strains of Cunnighamela sp. and compare it with mammalian model i.e. rat liver microsomes. LC–MS was used for analysis and prediction of the chemical structure of the metabolites in the sample.
Materials and methodsChemicalsBromhexine was received as a gift sample from Ven Petrochem & Pharma limited (Mumbai, India). NADPH, peptone, yeast extract, potato dextrose agar were obtained from Himedia (Mumbai, India). Methanol, ethyl acetate, triethylamine and acetonitrile were obtained from Loba chemie. All chemicals were of analytical grade or higher. All the three strains of Cunninghamella sp. were procured from National Collection of Industrial Microorganisms, Pune. Rat liver microsomes were obtained from Sigma Aldrich, Mumbai.
Characterization of Cunninghamella sp.Microscopic characterization of Cunninghamella elegans (NCIM 689), C. echinulata (NCIM 691) and C. blakesleeana (NCIM 687) was performed by slide culture preparation method.16
Fermentation and extraction of metabolitesAll the cultures were maintained on their respective agar slants at 4°C. They were grown on potato dextrose agar media. The medium used for fermentation was glucose – 20g, peptone – 5g, yeast extract – 5g, potassium phosphate – 5g, NaCl – 5g in 100mL distilled water.8 Fermentation for microbial metabolism was carried out in 250mL Erlenmeyer flask containing 50mL medium. The culture flasks were incubated in an orbital shaker, operating at 220rpm at 37°C for 24h. After 24h, 1mL from this culture flask was inoculated in another flask containing 20mL broth. The entire platform of orbital shaker moved in a circular orbit to provide good aeration and mixing which is better than the rotary shakers spinning. After 24h incubation, the drug substrate (bromhexine) 20μg/mL was added and kept in orbital shaker for 3, 5 and 7 days. The study included the substrate control to which substrate was added and incubated without microorganism. Culture control were maintained in which the microorganisms were grown under identical conditions without the substrate and solvent control with solvent methanol to check its effect on growth of culture. It was done to test whether bromhexine would be chemically decomposed or transformed during the incubation period.17 The fermentation batches were carried out in triplicates.
Extraction of metabolitesThe incubated medium was centrifuged at 3000rpm for 15min at 37°C. A clear supernatant liquid was collected, 8mL of which was extracted with 5mL of ethyl acetate. The upper organic layer was separated and evaporated at 72°C to concentrate the product. For HPLC analysis, it was reconstituted with 0.5mL of mobile phase and centrifuged at 13,000rpm for 10min. Aliquots (25μL each) were injected into the HPLC system for analysis. Controls were also prepared to provide suitable blanks.8,18
Inhibition studiesThe inhibition studies were carried out to confirm the involvement of specific CYP450 isoenzyme in bromhexine metabolism. One sample and four controls were used. These include a substrate control, inhibitor control and inhibitor and substrate control to check the interference of substrate, inhibitor alone and in combination with the media. Culture control comprised fermentation blanks in which the microorganisms were grown under identical conditions without the substrate. The test was performed in two stages; in the first stage, drug control, culture control and inhibitor control were incubated in orbital shaker for 72h. Inhibitor and substrate control and samples were incubated under the same conditions for 24h by adding 20μg/mL of inhibitor. In second stage, i.e., after 24h of incubation 0.5mL of drug solution was added to the inhibitor and substrate control and to the sample for a further 48h incubation. The contents of the flask were extracted and analyzed by HPLC to determine the extent of inhibition of metabolism. The experiments were carried out in culture flasks (250mL), each containing 50mL of broth, and incubated in a rotary shaker operated under similar conditions. The substrate (bromhexine), CYP3A4 inhibitor clarithromycin stock solutions, were prepared separately by dissolving 20mg in 10mL of methanol and acetone respectively.17,19 Methanol has been reported only to inhibit CYP2C9 and CYP2E1 not CYP3A4 activities.20 Thus methanol was considered safe for this study as solvents may inhibit the CYP enzyme's activity.
High performance thin layer chromatography (HPTLC)Extracted metabolites were characterized by thin layer chromatography. Merck Silica gel F254 plates were used as stationary phase. A semi polar solvent and glacial acetic acid was then tried to optimize the method. Since presence of primary amines cause an improper streak, glacial acetic acid was added for proper elution of the spots.21 Metabolite samples were applied as bands of 6mm width with a 100μL sample syringe on aluminum plates precoated with silica gel GF254 (20×10cm), using auto sampler. A constant application rate of 10μL/s was used, and the space between bands was 12mm. The slit dimension was 6.0×0.3μm and the scanning speed was 20mm/s. The mobile phase comprised glacial acetic acid:water:n butanol (17:17:66). Linear ascending development was carried out in a glass chamber saturated with the mobile phase. Development of the plates was left till the mobile phase migrated 9.5cm. Following the development, the plates were air dried, spots were visualized under UV lamp at 254nm and densitometric scanning was performed using DESAGA GAMBLR Scanner in the reflectance-absorbance mode at 270nm and operated by PROQUANT software. The radiation source was deuterium lamp.
Fourier transform-infrared spectroscopy of metabolites (FT-IR)Metabolite extracts were sandwiched between two plates of a potassium bromide salt. The plates were transparent to the infrared light and did not introduce any lines onto the spectra. Spectra obtained from the metabolites of the three strains of fungi were used to predict the metabolites produced.21
HPLC method development for metabolitesHigh performance liquid chromatography was carried out using Kromasil C18 column – 250×4.6mm stainless steel column. Various mobile phase concentrations were used for proper elution and separation of peaks. Elution was detected by UV detector at wavelength of 248nm. Injection volume was set at 20μL. Flow rate was adjusted to 1mL/min. Mobile phase concentrations used were acetonitrile:phosphate buffer (33:67) at pH 7.4 and acetonitrile:orthophosphoric acid buffer (85:15) at pH 7.4. The generation of microsomal metabolites were analyzed using a Perkin Elmer HPLC system equipped with a UV detector. The HPLC system was equipped with a degasser and an auto injector, and data was collected and analyzed using Totalchrom navigator software.21
LC–MSThe metabolites produced in the extracts of Cunninghamella sp. by HPLC analysis were further analyzed by mass spectrometry. LC–MS analysis was carried out with an ion trap mass spectrometer (TDM labs, Mumbai) in both positive and negative electron spray ionization (ESI) source system. The interface was adjusted to the following conditions: ion mode, positive and negative: spray voltage, 3.5kV: capillary temperature, 325°C; and auxiliary gas (nitrogen), 40psi.
Microsomal metabolismBromhexine metabolism mediated by RLM was tested in vitro. These assays were performed with an NADPH system in 100mM potassium phosphate buffer (pH 7.4). After the substrate was preincubated at 37°C for 5min, the enzymatic reactions were initiated by the addition of RLM with 20μg/mL protein content and incubated at 37°C for time period of 0, 15, 30 and 60min. Assay reactions were terminated by addition of an equal volume (50μL) of acetonitrile. After centrifugation at 15,000rpm for 5min, the supernatant was collected for metabolite characterization using an HPLC system. Incubated vials were capped to prevent loss of substrate due to volatility. For controls, each substrate was incubated in the same buffer system containing RLM without the NADPH-generating system.19,22
Stability studiesStability profile of metabolic extracts was tested by storage at −20°C for time periods of 30, 45, 60 and 90 days. Samples collected at different time points were analyzed by HPLC for any change in the production of metabolites.
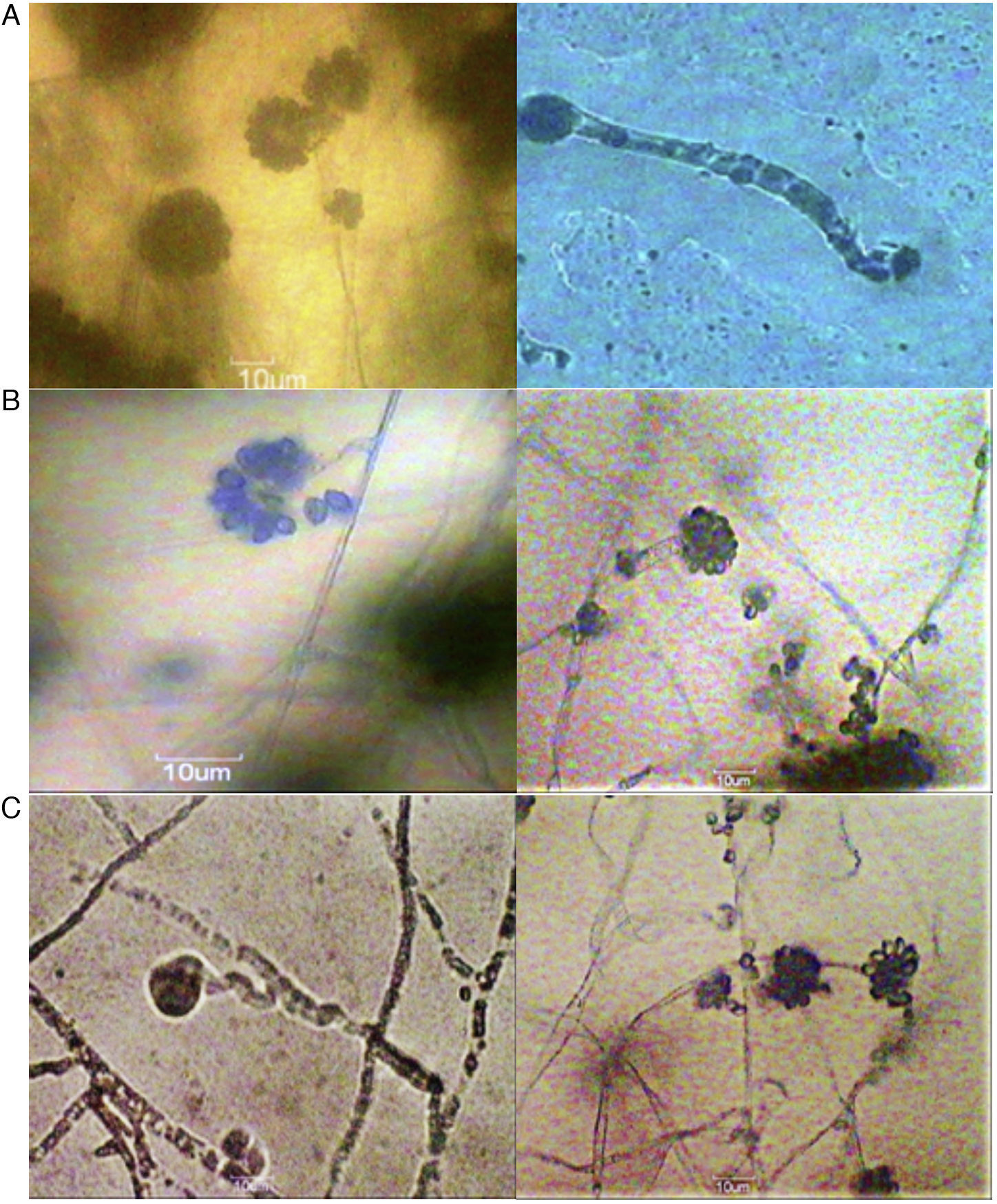
Results and discussionsCharacterization of Cunninghamella sp.The genus Cunninghamella was characterized by white to gray, rapidly growing colonies, producing erect, straight, branching sporangiophores. Initially colonies appeared white, turning later dark gray and powdery with sporangiola development. These sporangiophores ended in globose or pyriform-shaped vesicles from which several one-celled, globose to ovoid, echinulate or smooth-walled sporangiola developed on swollen pentacles (Fig. 2).
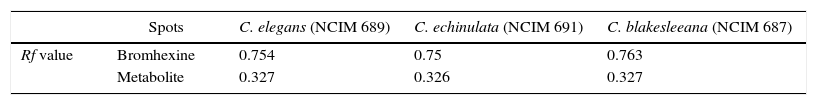
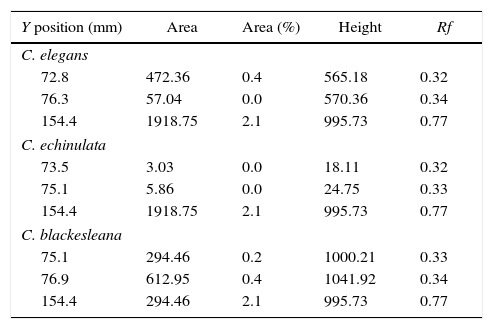
High performance thin layer chromatography of metabolites (HPTLC)After carrying out the biotransformation process, characterization of generated metabolites and bromhexine was done using HPTLC. The solvent system of n-butanol:glacial acetic acid:water (66:17:17) was found to be the optimum system for the separation of metabolites. All the three strains of Cunninghamella showed three spots in HPTLC and two in TLC. Proper elution was achieved and the Rf of metabolites and drug was found to be similar in all the three strains. Bromhexine spot was visible at Rf 0.75 and other two were metabolite spots (Table 1).
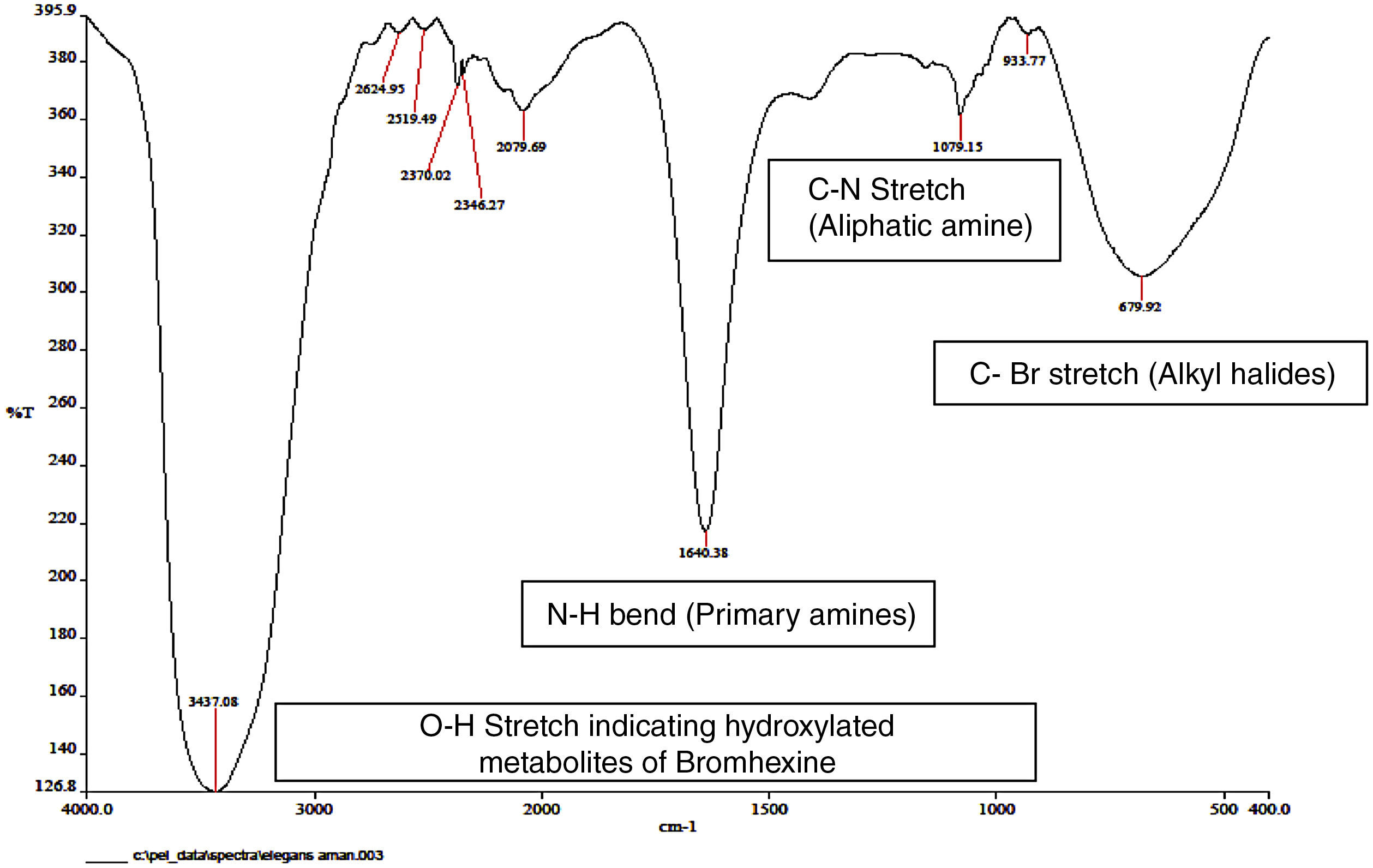
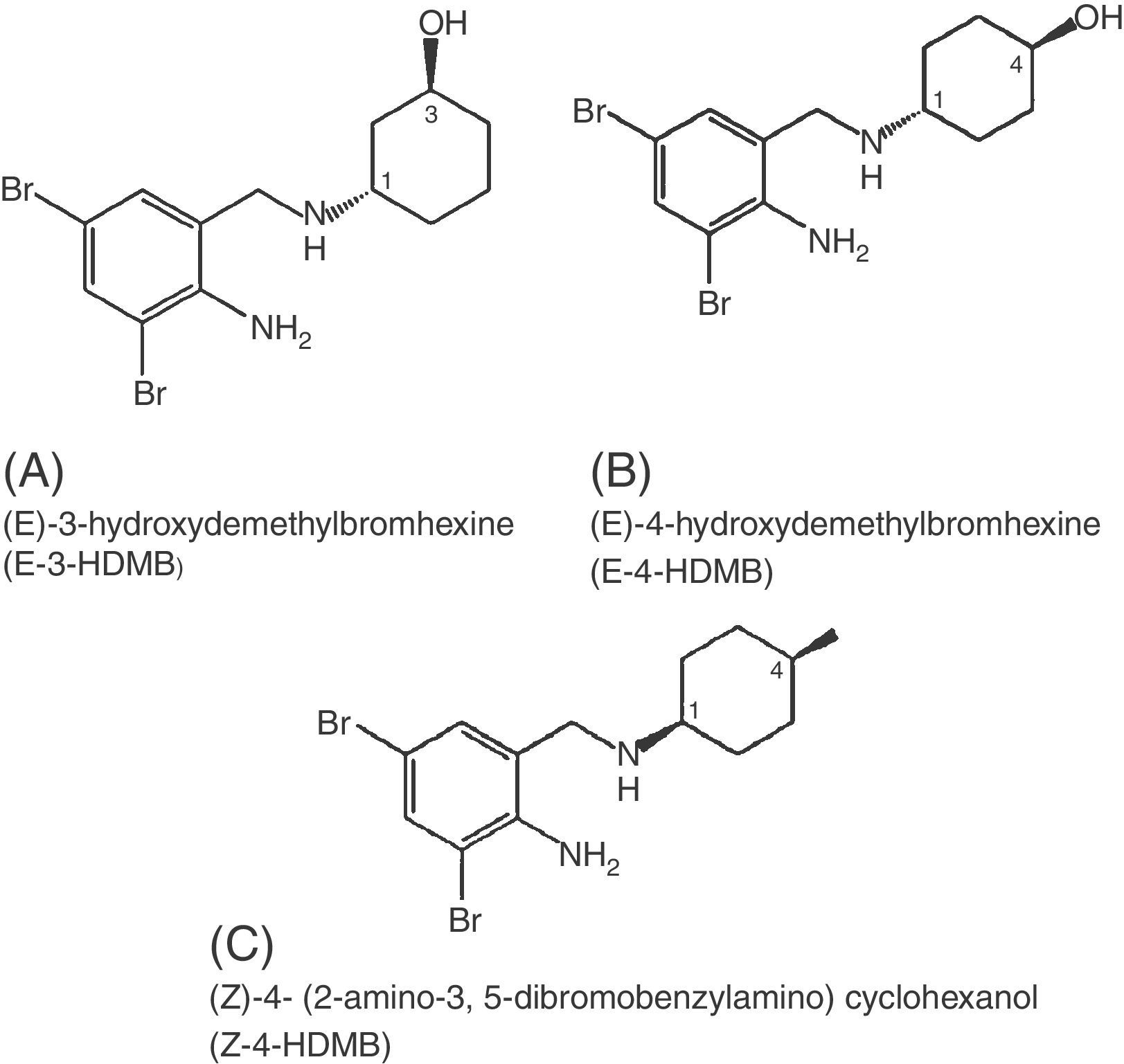
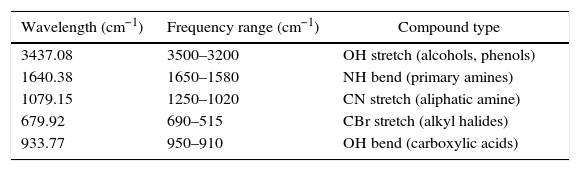
FT-IR spectroscopy of metabolitesInfrared spectroscopy of all the three fungal metabolite extracts were done and the results obtained are shown in Table 2 and Fig. 3. KBr was used as an internal standard. Metabolite structures were predicted as hydroxy demethylated bromhexine derivatives (Fig. 4) based on the wavelengths and reports in literature.13
Interpretation of IR spectra of Bromhexine metabolites.
| Wavelength (cm−1) | Frequency range (cm−1) | Compound type |
|---|---|---|
| 3437.08 | 3500–3200 | OH stretch (alcohols, phenols) |
| 1640.38 | 1650–1580 | NH bend (primary amines) |
| 1079.15 | 1250–1020 | CN stretch (aliphatic amine) |
| 679.92 | 690–515 | CBr stretch (alkyl halides) |
| 933.77 | 950–910 | OH bend (carboxylic acids) |
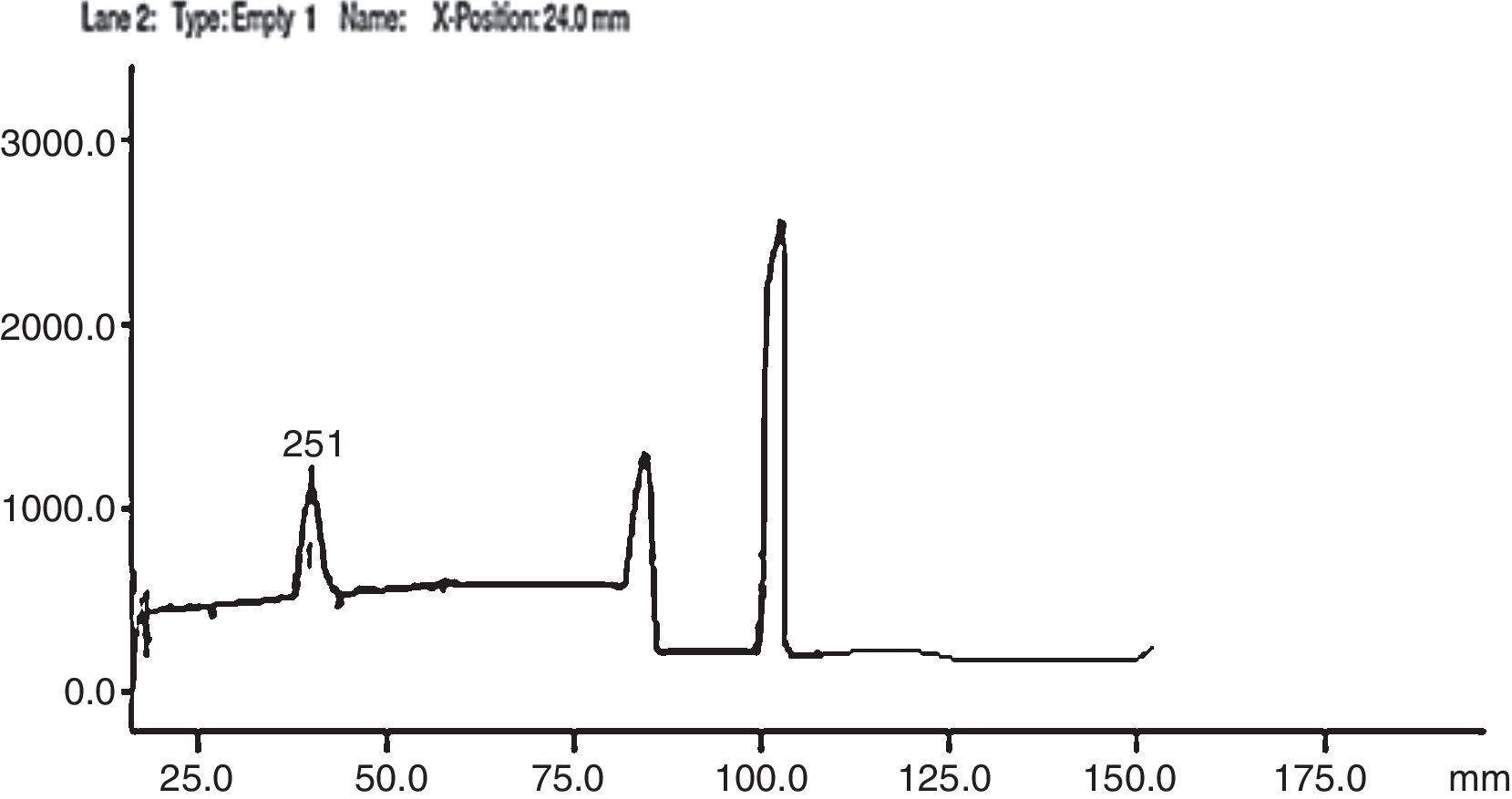
HPTLC scanning was done in the wavelength range of 200–800nm. λmax was detected at 254nm as reported in Indian Pharmacopeia.21 Saturation of the glass chamber was done with the mobile phase for 45min. The plates were developed for a distance of 9.5cm. Three bands were observed in all the three fungal extracts and appeared as distinct peaks in the chromatogram (Fig. 5). Based on peak area and retention time, bromhexine conversion to polar metabolites was observed. C. echinulata produced similar pattern of metabolites as C. elegans but there was downward trend in peak area observed showing less metabolite concentration and thus less conversion when compared to C. elegans (Table 3). Peak areas were greater in C. blackesleana and the results confirmed the metabolite generation by all the three strains of Cunninghamella by HPTLC method.
HPTLC peaks and their Rf values.
| Y position (mm) | Area | Area (%) | Height | Rf |
|---|---|---|---|---|
| C. elegans | ||||
| 72.8 | 472.36 | 0.4 | 565.18 | 0.32 |
| 76.3 | 57.04 | 0.0 | 570.36 | 0.34 |
| 154.4 | 1918.75 | 2.1 | 995.73 | 0.77 |
| C. echinulata | ||||
| 73.5 | 3.03 | 0.0 | 18.11 | 0.32 |
| 75.1 | 5.86 | 0.0 | 24.75 | 0.33 |
| 154.4 | 1918.75 | 2.1 | 995.73 | 0.77 |
| C. blackesleana | ||||
| 75.1 | 294.46 | 0.2 | 1000.21 | 0.33 |
| 76.9 | 612.95 | 0.4 | 1041.92 | 0.34 |
| 154.4 | 294.46 | 2.1 | 995.73 | 0.77 |
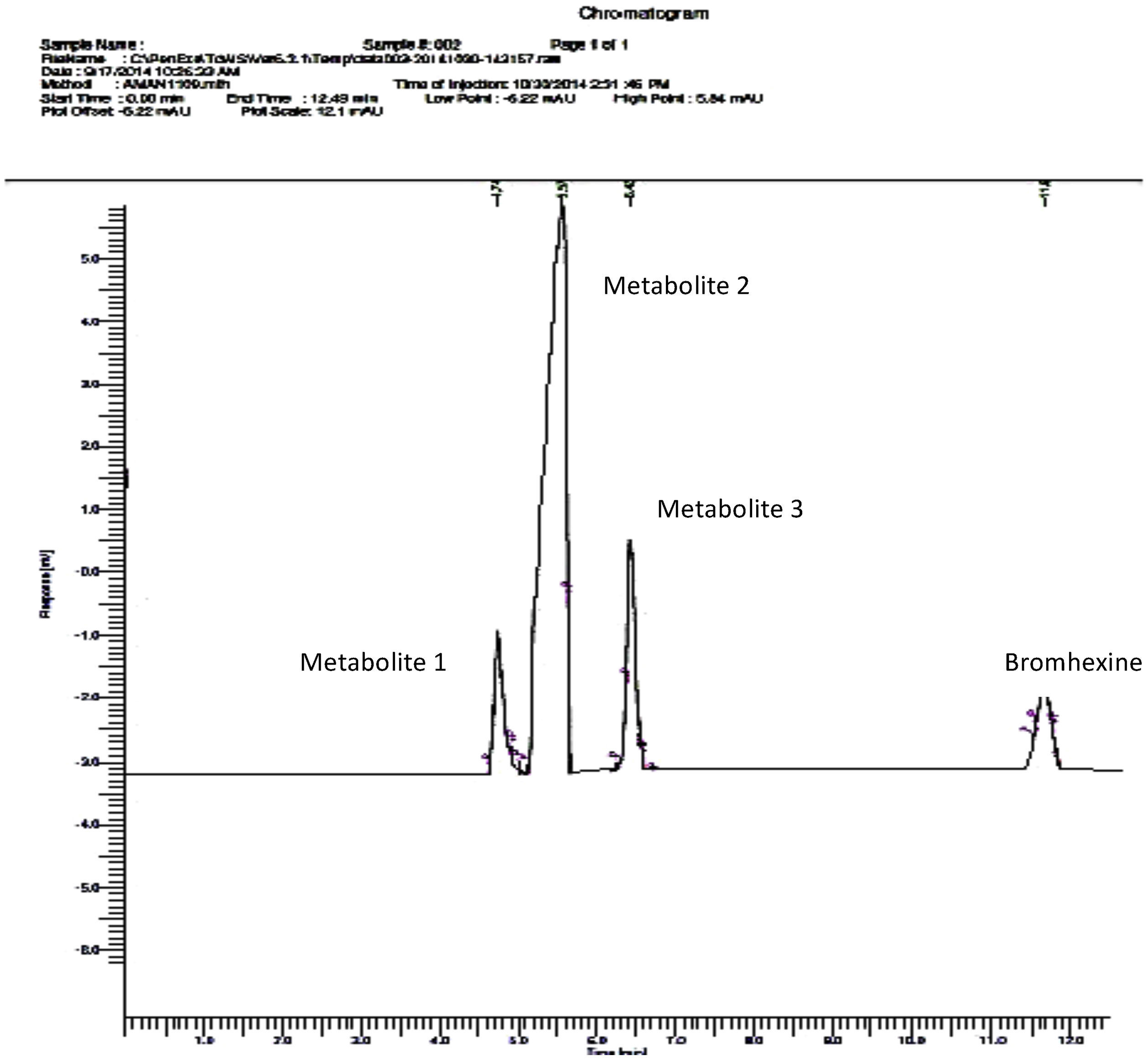
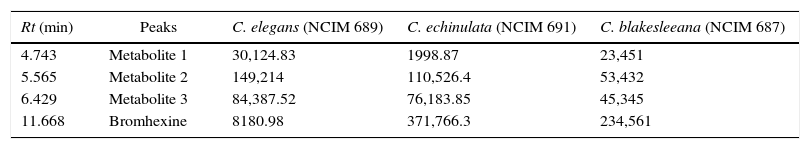
Overlapping and improper peak separation was observed with mobile phase of acetonitrile 0.01mol/L:phosphate buffer (containing 0.2% triethylamine) (33:67) in the study reported by Lad et al.23 In our study also, when polarity was decreased and acetonitrile concentration was adjusted to 85%, better elution was observed. Thus, acetonitrile 0.01mol/L:0.1% orthophosphoric acid buffer (85:15) (pH 7.4) was used as optimized mobile phase for better elution of metabolites. In conventional C18 column the silica backbone contains free silanol group SiOH. After ionization, it produces ionic retention mechanism where positive molecule species like amines, NH2 group gets attracted and retain longer on the column bed producing a hideous tailing chromatogram peak. So, to avoid this tailing effect, triethylamine was used to mask free silanol groups on the HPLC column and therefore to improve peak shape of the analytes. Moreover it served both the purpose of pH adjustment and peak shape improvement. It acts as a competing base, saturating available silanols, thus reducing the potential for tailing.24 If no triethylamine was added, bromhexine would elute as a distorted and broadened peak. With increase in triethylamine content, both bromhexine and its metabolites exhibited change in retention because of small pH change. 0.1% triethylamine was selected for minimal retention time with better peak shapes of bromhexine and its metabolites. The flow rate was 1.0mL/min. Metabolites were separated by a reversed phase C18 column (250×4.6mm) and detected using a UV detector at 254nm. 50μL of sample was injected into the system. Based on the peak area % of different strains, C. elegans showed maximum metabolite peak area % and minimum bromhexine substrate area at Rt 11.668 indicating maximum conversion by this strain (Fig. 6). C. echinulata showed second best conversion followed by C. blakesleeana (Table 4).
HPLC retention time Rt (min) and the peak area for the three fungal metabolites extract.
| Rt (min) | Peaks | C. elegans (NCIM 689) | C. echinulata (NCIM 691) | C. blakesleeana (NCIM 687) |
|---|---|---|---|---|
| 4.743 | Metabolite 1 | 30,124.83 | 1998.87 | 23,451 |
| 5.565 | Metabolite 2 | 149,214 | 110,526.4 | 53,432 |
| 6.429 | Metabolite 3 | 84,387.52 | 76,183.85 | 45,345 |
| 11.668 | Bromhexine | 8180.98 | 371,766.3 | 234,561 |
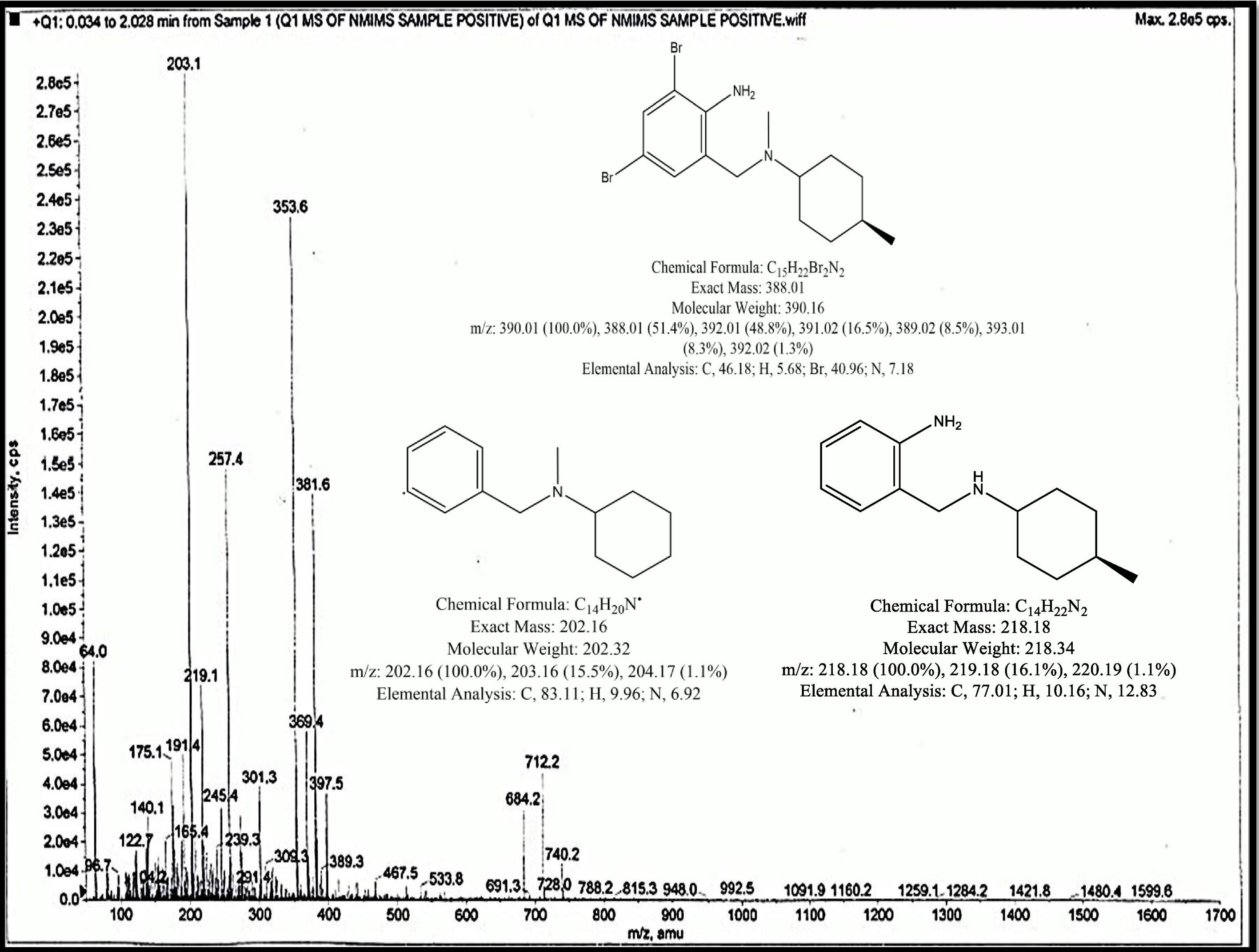
LC–MS spectra of the fungal metabolite extract showed the protonated form of bromhexine [M+H+4]+ at m/z 381.6 in positive ionization mode as earlier reported.13 Major characteristic fragments were observed in positive ionization mode than negative mode. It also represented the major metabolite of bromhexine named ambroxol at m/z 389. Because of the presence of two bromine atoms in the structure of bromhexine, the soft ionization process in the ESI source produced the protonated molecules [M+H+4]+ at m/z 381.6. Under the experimental conditions, the product ion mass spectrum of [M+H+2]+ of bromhexine showed intense fragments at m/z 257.4 and m/z 122.7, formed by the cleavage of an amide bond. m/z at 219.3, 389 and 203.1 occurred due to dealkylation, hydroxylation of bromhexine and debromination of the metabolites in positive ionization respectively (Fig. 7).
Microsomal metabolismBromhexine was incubated with rat liver microsomes in the presence of NADPH. When rat liver microsomes were used, the HPLC profile showed two new peaks, which were not detectable when NADPH was omitted from the reaction mixture. In a typical HPLC profile, bromhexine constituted a prominent absorption peak eluting at 12min, while the three bromhexine metabolites eluted at 4, 10 and 14min. Their retention times were corresponding to those of the microbial metabolites generated by Cunnighamela sp. The rate of formation of metabolite was linear up to 60min. At the end of 60th minute, maximum conversion was observed with better peak response of 30.05%, 21.06%, 1.34%, and 47.66%. Area % of metabolites peak were comparable to microbial metabolites. Area % of metabolite peak produced by C. elegans was found to be greater than RLM's.
Stability studies of metabolitesMetabolites were found to be stable after storage for 30 days, since all the peaks with the same retention time and peak area were observed with no drastic decrease in the peak area %.
Inhibition studiesWe investigated the role of inhibitor drug clarithromycin to see the substrate inhibition by CYP3A4 for bromhexine metabolites. To confirm the involvement of CYP3A4 in the formation of metabolite, RLM's and microbial cultures were incubated with clarithromycin, a potent CYP3A4 inhibitor. The formation of metabolites was completely inhibited in the presence of 20μg/mL clarithromycin. In case of bromhexine, the elution profile of its metabolites was permuted by clarithromycin, indicating role of CYP3A inhibitor in the metabolism of bromhexine. Cunninghamella sp. was concluded to metabolize bromhexine by CYP3A4 as reported in mammals. There was no metabolite generation when C. elegans was incubated with clarithromycin. The peaks produced were negligible. Similar trend was observed in C. echinulata and C. blackesleana sp. The results confirmed the role of CYP3A4 in bromhexine metabolism and also the presence of these enzymes in Cunninghamella sp.
ConclusionThe generation of ambroxol in human plasma metabolism has been known, but there are no studies done for its production by fungal metabolism. Ambroxol and other metabolites generation by Cunninghamella sp. has not been previously reported. Furthermore, CYP3A4 has been observed as the dominant isoform for bromhexine metabolism. We have determined the major biotransformation products of the mucolytic drug bromhexine by fungus C. elegans, C. echinulata and C. blackesleana. The work showed that ambroxol (demethylated bromhexine), a predominant metabolite of bromhexine was produced by Cunninghamella sp. In one of the earlier studies, Kopitar et al. (1973) reported that there is difference in metabolism and excretion of bromhexine in mice, rats, rabbits, dogs and humans; and the pattern of rabbit is most similar to humans, while least similarity is reported in rats.25 In our study, we have found similarity in the bromhexine metabolism in Cunninghamella and RLMs, and confirm that fungi has the comparable potential for metabolizing bromhexine with rat liver microsomes.
Four metabolites have been reported in human plasma, among which (E)-4-hydroxydemethylbromhexine (E-4-HDMB; ambroxol) and (E)-3-hydroxydemethylbromhexine (E-3-HDMB) are major and (Z)-4-hydroxydemethylbromhexine and (Z)-3-hydroxydemethylbromhexine are minor metabolites.13 In our result intense peaks at m/z 257.4, 122.7, 219.3, 389 and 203.1 occurred due to dealkylation, hydroxylation of bromhexine and debromination of the metabolites respectively. In conclusion, the microbial transformation of bromhexine showed the higher potential of Cunninghamella species to produce metabolites of bromhexine. Therefore, these microbial models can be used to predict potential routes of mammalian metabolism of drug candidates in the early phase of drug discovery and development. Earlier studies by Rydevik (2014), showed the detection of glutathione, glutathione ethyl-ester and cysteine conjugates and other phase II reactions concluding the fungus use in the drug development process to identify possible drug toxicity at an early stage.26 Further toxic metabolites of certain drugs can be predicted and tested by microbial model of metabolism. In addition to this, the microbes can be further used for production of metabolites by fermentative scale up and purification. This process can be used for a cost effective method for both known and unknown metabolite production and characterization.
Conflicts of interestThe authors declare no conflicts of interest.
This research was financially supported by the SVKM'S NMIMS University as an In-house project.