Psittacine birds have been identified as reservoirs of diarrheagenic Escherichia coli, a subset of pathogens associated with mortality of children in tropical countries. The role of other orders of birds as source of infection is unclear. The aim of this study was to perform the molecular diagnosis of infection with diarrheagenic E. coli in 10 different orders of captive wild birds in the state of São Paulo, Brazil. Fecal samples were analyzed from 516 birds belonging to 10 orders: Accipitriformes, Anseriformes, Columbiformes, Falconiformes, Galliformes, Passeriformes, Pelecaniformes, Piciformes, Psittaciformes and Strigiformes. After isolation, 401 E. coli strains were subjected to multiplex PCR system with amplification of genes eae and bfp (EPEC), stx1 and stx2 for STEC. The results of these tests revealed 23/401 (5.74%) positive strains for eae gene, 16/401 positive strains for the bfp gene (3.99%) and 3/401 positive for stx2 gene (0.75%) distributed among the orders of Psittaciformes, Strigiformes and Columbiformes. None of strains were positive for stx1 gene. These data reveal the infection by STEC, typical and atypical EPEC in captive birds. The frequency of these pathotypes is low and restricted to few orders, but the data suggest the potential public health risk that these birds represent as reservoirs of diarrheagenic E. coli.
Many zoonoses are associated with ecological imbalances as a result of deforestation, human population expansion, changes in agricultural practices and encroachment on wildlife habitats.1 Urban areas represent impacted environment and wild animals are maintained in captivity, living in zoos. Captive wildlife animals are very susceptible to opportunistic diseases and they may act as reservoir of pathogenic bacteria.2,3
Escherichia coli can be considered the most prevalent opportunistic enterobacteria in captive animals and were associated with systemic disease in birds.4 Airsaculitis and sepsis are often caused by avian pathogenic Escherichia coli (APEC), which are considered as Extraintestinal pathogenic E. coli (ExPEC) pathotype.5 The pathogenesis of enteritis by E. coli in birds is still unclear, but the presence of diarrheagenic strains may represent a public health risk. Shiga toxin-producing E. coli (STEC) and enteropathogenic E. coli (EPEC) represent two of at least six pathotypes of human diarrheagenic E. coli that affect birds and may be considered zoonotic pathogens.6,7
The EPEC pathotype leads to a high child mortality rate in developing countries. Diarrhea is a consequence of loss of intestinal microvillus, after bacterial adherence on enterocytes, with ensuing attaching and effacing (A/E) lesions.8 The AE lesion depends on intimin, an outer membrane protein, codified by the eae gene, which is present in the pathogenicity island termed the locus of enterocyte effacement (LEE).9 The identification of typical EPEC strains (tEPEC) is performed by molecular detection of the eae and bfp genes (“bundle-forming pili” encoded by the EAF plasmid). The EPEC are regarded as atypical (aEPEC) in case of absence of the EAF plasmid, making it eae+ and bfp−.10
The attaching and effacing genes on the LEE may be also present in STEC, but Shiga toxin production is considered the most relevant virulence factor of this diarrheagenic E.coli patothype.8,11 Shiga toxins can be classified into two types, Stx1 (subtypes a, c and d) and Stx2 (subtypes a to g). Both toxins are encoded on prophages that are integrated into the chromosome and lead to inhibition of protein synthesis, causing cell death.8
In humans, STEC infection causes hemorrhagic colitis following injury of the intestinal epithelium, induced by Shiga-toxin production. Stx-2 toxin is more toxic than Stx-1, and is often associated with hemolytic uremic syndrome.8 The transmission routes of STEC include ingestion of contaminated food or water and contact with infected companion animals (dogs, cats and birds).7,8,11
The aim of this study was to search for the presence of EPEC and STEC isolates in captive birds from different orders located at zoos from São Paulo, state, Brazil.
Material and methodsBirdsThis project was approved by the Ethics Committee of São Paulo University (2984230514) and authorized for scientific purposes (SISBIO 43541-1). We examined a total of 516 fecal samples isolated from captive birds belonging to 10 orders (including 70 species): Accipitriformes (hawk, n=14), Galliformes (guan and curassow, n=50), Anseriformes (duck and goose, n=80), Psittaciformes (macaw, parrot and parakeet, n=99), Passeriformes (canary and thrush, n=88), Falconiformes (falcon, n=46), Strigiformes (owl, n=48), Columbiformes (pigeons, n=72); Piciformes (toucan and aracari, n=10) Pelecaniformes (pelican and egret, n=9).
The samples were collected from September 2013 to June 2015, in two municipal zoos located in São Paulo State, Brazil. Fecal swabs were seeded in Amies transport media and sent to the laboratory, under refrigerated conditions.
Culture and Identification of E. coliThe fecal samples were enriched in brain heart infusion broth, seeded on MacConkey agar, and incubated at 37°C for 24h. Bacteria were identified by biochemical tests, using an Enterokit (Probac® – São Paulo, Brazil).
PCR Amplification for virulence genesSearch for virulence genes in the diarrheagenic E. coli isolates performed by PCR for amplification of eae (454bp), bfp (550bp), stx1 (349bp) and stx2 (110bp) genes, according to the method described by Costa et al. (2010).12 The following strains were used as control of the PCR: E. coli DH5α (negative control); O157:H7 (STEC positive control) and O55:H7 (EPEC positive control).
The DNA extraction was performed as described by Boom et al. (1990).13 The amplification mixture consisted of Tris-HCl buffer (pH 8.3) 10mM, MgCl2, deoxynucleotide triphosphates 200mM, pairs of primers, Taq DNA polymerase 0.5U, and ultrapure water autoclaved in a final volume of 25μl. Amplified products were separated in 1.5% agarose gel and examined after stained with BlueGreen® (LGC Biotecnologia, São Paulo, Brazil). A 100bp DNA ladder (LGC Biotecnologia, São Paulo, Brazil) was used as a molecular size marker.
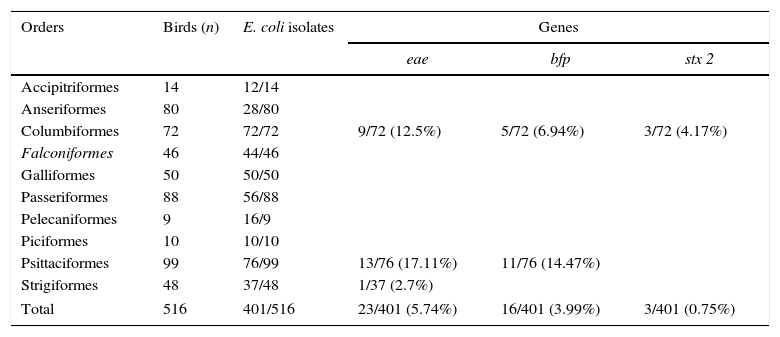
ResultsA total of 401 isolates were identified as E. coli. After PCR investigation, 23/401 isolates were positive for eae, 16/401 positive for bfp and 3/401 positive for stx2 genes (Table 1). None of strains (0/401) were positive for stx1 gene.
Isolation of E. coli and virulence genes distributed according to the orders of captive wild birds. São Paulo, 2013–2015.
| Orders | Birds (n) | E. coli isolates | Genes | ||
|---|---|---|---|---|---|
| eae | bfp | stx 2 | |||
| Accipitriformes | 14 | 12/14 | |||
| Anseriformes | 80 | 28/80 | |||
| Columbiformes | 72 | 72/72 | 9/72 (12.5%) | 5/72 (6.94%) | 3/72 (4.17%) |
| Falconiformes | 46 | 44/46 | |||
| Galliformes | 50 | 50/50 | |||
| Passeriformes | 88 | 56/88 | |||
| Pelecaniformes | 9 | 16/9 | |||
| Piciformes | 10 | 10/10 | |||
| Psittaciformes | 99 | 76/99 | 13/76 (17.11%) | 11/76 (14.47%) | |
| Strigiformes | 48 | 37/48 | 1/37 (2.7%) | ||
| Total | 516 | 401/516 | 23/401 (5.74%) | 16/401 (3.99%) | 3/401 (0.75%) |
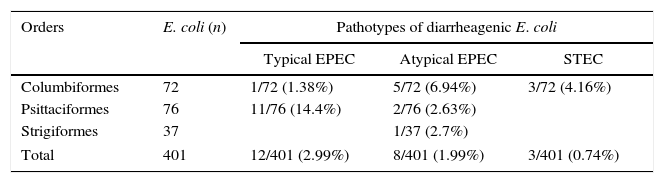
The pathotype classification of the isolates is presented in Table 2. These results showed that atypical EPEC were detected in 3/10 orders, including Psittaciformes (2/99 birds), Columbiformes (2/72 birds) and Strigiformes (1/48 birds). Typical EPEC were detected in 2/10 orders, including Psittaciformes (11/99 birds) and Columbiformes (4/72 birds). STEC was detected only in Columbiformes, present in 3/72 birds.
Pathotypes of diarrheagenic E. coli distributed according to the orders of captive wild birds. São Paulo, 2013–2015.
| Orders | E. coli (n) | Pathotypes of diarrheagenic E. coli | ||
|---|---|---|---|---|
| Typical EPEC | Atypical EPEC | STEC | ||
| Columbiformes | 72 | 1/72 (1.38%) | 5/72 (6.94%) | 3/72 (4.16%) |
| Psittaciformes | 76 | 11/76 (14.4%) | 2/76 (2.63%) | |
| Strigiformes | 37 | 1/37 (2.7%) | ||
| Total | 401 | 12/401 (2.99%) | 8/401 (1.99%) | 3/401 (0.74%) |
The total prevalence of diarrheagenic E. coli strains was 23/401 (5.74%), which included 13/76 (17.03%) from Psittaciformes, 9/72 (12.48%) from Columbiformes and 1/37 (2.7%) from Strigiformes.
DiscussionBrazil has great wildlife biodiversity. A wide range of birds species are also kept in zoos for entertainment, education, search and conservation. Our study analyzed 10 orders with 70 species of birds for the presence of diarrheagenic E. coli. Unfortunately, due to the small number of birds in some orders, a fair sampling was somewhat compromised. Our results demonstrate that molecular techniques were useful for diagnosis of the diarrheagenic E. coli pathotypes, identifying captive birds infected by typical or atypical EPEC and STEC.
The results showed that EPEC were found in 3/10 orders, including Psittaciformes (2/99 birds), Columbiformes (2/72 birds) and Strigiformes (1/48 birds). Likewise, Kobayashi et al. (2009) evaluated the prevalence of eae- and stx-positive E. coli strains in 447 wild birds belonging to 62 species in Tokyo.14 Eae-positive strains were found in 7/10 orders: Columbiformes, Passeriformes, Anseriformes, Ciconiformes, Procellariformes, Pelecaniformes and Galliformes. However, the prevalence of eae-positive strains in Tokyo was 25% (11/447), higher than the prevalence of this study (5.74%). Psittaciformes were not included in a Tokyo survey, but Passeriformes, Columbiformes and Pelecaniformes were implicated as a reservoir of EPEC.14 In Brazil, none of the Passeriformes, Pelecaniformes or Anseriformes investigated were infected by EPEC. However, we found one atypical EPEC in Strigiformes (1/37 owl). To our knowledge, this is the first report of infection by EPEC in owls.
Relative analyses showed that Psittaciformes is the most prevalent order of birds positively infected by EPEC (Table 2), with 14.4% of typical EPEC and 2.63% of atypical EPEC. The EPEC infection of psittacine birds in Brazil was reported previously in parrots (Amazona aestiva, Amazona amazonica) and macaws (Anodorhynchus leari, Guarouba guarouba) with prevalence ranging from 2.27% in free-ranging birds15 to 6.5% for captive psittacine birds.16
Marietto-Gonçalves et al. (2011) investigated swabs from 86 psittacidaes recovered from illegal wildlife trade in Brazil and found only one strain (1/86 – 1.1%) classified as typical EPEC, isolated from a blue-fronted parrot.17 We believe that the high prevalence reported in our study (11.11%) is related to zoo enclosures that allow intense contact between birds, mammals and park visitors. Bacterial diversity was reported previously to be significantly lower in wild parrots and the composition of cloacal bacterial microbiota might undergo significant changes in captive birds if they are overexposed to contact with mammals.18
Farooq et al. (2009) found a high prevalence of atypical EPEC (15.56%) from avian species in India.6 The frequency was greater in farmed animals (chicken and duck – 27/112) than in pigeons (6/100), but the authors still believe that pigeons act as an infectious source for commercial poultry. Our survey also highlights the role of Columbifomes as a reservoir of EPEC and STEC. We detected 6/72 EPEC and 3/72 STEC strains in feral pigeons (Columba livia). This data are similar to those reported by Kobayashi et al. (2009), with 5/67 EPEC and 2/67 STEC strains in Tokyo.14
Feral pigeons are synanthropic birds. In zoos, these birds invade the enclosures looking for water and food, and transmit diseases or even acquire pathogens from animals belonging to other classes, such as mammals and reptiles. The zoonotic risk associated with EPEC infection in pigeons was first documented by Silva et al. (2009) in Brazil, reporting 3.3% prevalence of infected pigeons in urban areas.19 Population control measures in urban environments are very difficult due to the absence of natural predators. The presence of these birds in some habitats has been associated with transmission of many zoonoses such as chlamydiosis, salmonellosis and campylobacteriosis.20
Sacristan et al. (2014) reported that feral pigeons from Spain were infected with atypical EPEC (8%).21 The frequency of eae gene in urban pigeons was 6% and 4% in rural species. The authors highlighted the public health risks associated with antibiotic resistance, because some strains presented class I integrons containing genes cassetes encoding for antibiotic resistance.
We believe that the colonization of birds may vary according to the susceptibility of the species, with influence of diet, microflora and management. Apparently, the orders of Galliformes and Anseriformes are less susceptible, and the reports of diarrheagenic E. coli in these birds are rare, even in the face of many management risk factors, such as access to lakes and water collection, contact with other animals in shared enclosures and difficulty in maintaining hygiene on dirt floors.
ConclusionThis study highlights the presence of diarrheagenic E. coli (EPEC and STEC) in captive Psittaciformes, Columbiformes and Strigiformes. The zoonotic potential of these strains may be investigated because of the sanitary impact on zoo bird collections, which are important for the “in situ” conservation of the species.
Conflicts of interestsThe authors wish to state that they have no competing interests with regard to the publication of this manuscript.
The authors are thankful to Quinzinho de Barros Zoo (Sorocaba, SP – Brazil) and Parque Estoril. Zoológico Municipal de São Bernardo do Campo for fecal samples donation.
This study was conducted with financial support from CNPq and FAPESP (2014/07837-6).