Role of microbes in bioremediation of oil spills has become inevitable owing to their eco friendly nature. This study focused on the isolation and characterization of bacterial strains with superior oil degrading potential from crude-oil contaminated soil. Three such bacterial strains were selected and subsequently identified by 16S rRNA gene sequence analysis as Corynebacterium aurimucosum, Acinetobacter baumannii and Microbacterium hydrocarbonoxydans respectively. The specific activity of catechol 1,2 dioxygenase (C12O) and catechol 2,3 dioxygenase (C23O) was determined in these three strains wherein the activity of C12O was more than that of C23O. Among the three strains, Microbacterium hydrocarbonoxydans exhibited superior crude oil degrading ability as evidenced by its superior growth rate in crude oil enriched medium and enhanced activity of dioxygenases. Also degradation of total petroleum hydrocarbon (TPH) in crude oil was higher with Microbacterium hydrocarbonoxydans. The three strains also produced biosurfactants of glycolipid nature as indicated d by biochemical, FTIR and GCMS analysis. These findings emphasize that such bacterial strains with superior oil degrading capacity may find their potential application in bioremediation of oil spills and conservation of marine and soil ecosystem.
Oil pollution is a perpetual problem that affects terrestrial as well as marine ecosystems. Oil spillage may arise either accidentally or operationally during production, transportation, storage, processing or when used at sea or on land.1 Crude oil is a multifarious mixture of many petroleum hydrocarbons of which polycyclic aromatic hydrocarbons (PAH) constitute a major fraction2 which invite greater attention owing to their toxic, mutagenic and carcinogenic nature.3 The recovery of spilled crude oil could be achieved by adopting physical and chemical methods but they can take care of only 10–15% of oil spillage. On the other hand, bioremediation by exploiting complex microbial communities can act as a self-driven, economical and eco-friendly method. Highly hazardous oily materials can easily be mineralized to harmless end products by introducing suitable microbial strains.4 Poly aromatic hydrocarbons (PAH) can act as carbon source for certain microbial communities in oil-polluted environment.5 Many bacterial species including Pseudomonas aeruginosa, Bacillus subtilis, Bacillus megaterium, Corynebacterium kutscheri, have been reported to possess oil degrading potential.6,7 The biochemical process of crude oil degradation by microbes involves the action of several enzymes including oxygenases, dehydrogenases and hydroxylases that fragment aromatic and aliphatic hydrocarbons. A major constraint in bio-degradation of oil is its hydrophobicity but biosurfactants produced by oil degrading bacteria facilitate uptake of hydrocarbons by the bacterial cells. Hence, those bacterial strains that possess the ability to produce biosurfactants along with enhanced oil degrading capacity are widely recommended for use in order to achieve fast degradation of crude oil.8
In this study, among the 12 bacterial isolates from oil contaminated soil samples, three strains which had the isolate designation of PA, PM and BM were selected owing to their superior oil degrading activity. These three strains were subsequently identified by 16S rRNA gene sequencing as Corynebacterium aurimucosum, Acinetobacter baumannii and Microbacterium hydrocarbonoxydans respectively. With these three strains, their ability to utilize crude oil as the sole carbon source, determination of activity of enzymes – Catechol 1,2 dioxygenase (C12O) and Catechol 2,3 dioxygenase (C23O) and characterization of biosurfactants produced by them were carried out. Additionally, the extent of degradation of aliphatic and aromatic compounds of total petroleum hydrocarbon (TPH) in crude oil by these strains was also analyzed.
Materials and methodsSampling and isolation of crude oil degrading bacterial strainsCrude oil and soil samples from crude oil contaminated sites were collected from Indian Oil Corporation Limited (IOCL), Chennai, India. For the isolation of crude oil degrading bacteria, 1g of soil sample was inoculated into 100mL of mineral salt medium (MSM pH 6.8±0.2) supplemented with 1% crude oil9 and incubated at room temperature for 1-week. From this, 1mL of active inoculum was again transferred to fresh MSM containing 1% crude oil and was used for the isolation of crude oil degrading bacteria by serial dilution technique followed by spread plate technique using nutrient agar (NA) plates (pH 7.4±0.2). The bacterial colonies were then cultured on fresh MSM agar plates supplemented with 1% (w/v) crude oil and those bacterial colonies grown were picked and preserved in liquid nutrient broth containing 70% glycerol at −80°C.
Determination of bacterial growth and activity of C12O and C23O enzymesThe ability of PA, PM and BM to utilize crude oil as carbon source was assessed by measuring their growth as colony forming units9 (log CFU) in MSM containing different concentrations (1, 2 and 3%) of crude oil (incubated at 30°C in an orbital shaker at 200rpm) for 13 days at 24h intervals. The enzyme activity of C12O and C23O were also determined in the cell free extracts of PA, PM and BM cultures at 24h intervals for 13 days. For this purpose, cells were harvested by centrifugation at 5000rpm (4°C), washed twice with 0.01M phosphate buffer (pH 7.0), subjected to sonication (15s) and the cell free extracts were collected by centrifugation at 20,000rpm for 25min (4°C). The activity of C12O was determined by detecting the levels of cis, cis-muconate10 and C23O based on the formation of 2-hydroxymuconic semialdehyde11 by UV–Vis spectrophotometer (Shimadzu, UV-Pharmaspec 1700). Protein concentration was determined by the Bradford method.12
Purification of C12OThe partial purification of C12O from the cell free extracts of PA, PM and BM was carried out by ammonium sulphate precipitation followed by ion exchange chromatography (DEAE cellulose). The cell free extract was treated with cold saturated (NH4)2SO4 solution (pH 7.5) to give 40, 60 and 80% saturation and the precipitates formed were collected by centrifugation at 10,000rpm for 20min (4°C). The pellets were dissolved in Tris–HCl buffer 50mM, (pH 7.5) and the resulting protein solutions were desalted by dialysis.13 The dialyzed protein sample was then loaded on to a DEAE cellulose column (10cm×2cm) that had been pre-equilibrated with 50mM Tris–HCl (pH 7.5). Elution of protein was carried out with 50mM Tris–HCl (pH 7.5) buffer containing NaCl in a linear gradient from 0.1 to 0.5M.14 The fractions collected were analyzed for enzyme activity and those fractions with highest activity were pooled and subjected to SDS PAGE analysis15 to determine the molecular weight and the purity of the C12O. Molecular weight marker (97.4–14.3kDa) (Merck bioscience) was used in this analysis.
Physicochemical and biochemical characterization of biosurfactantsTo characterize the biosurfactants produced by PA, PM and BM, the bacterial strains were inoculated individually in to MSM broth containing 1% crude oil and incubated at 30°C for 5 days in a shaker (200rpm). The cell free supernatant was collected by centrifugation at 10,000rpm for 20min and was subjected to physicochemical and biochemical characterization. The emulsification index (E24)16 and the oil displacement capacity17 of the cell free supernatant was determined. Qualitative analysis to detect glycolipids and lipoproteins in the cell free supernatants of PA, PM and BM was carried out respectively by Phenol sulfuric acid18 and Bradford Method.12 The sugar content was quantitatively determined by orcinol assay.19
Extraction and characterization of biosurfactants by FTIR and GCMS analysisThe biosurfactants from the cell free supernatants of PA, PM and BM were extracted by the acid precipitation method20 and the extracted biosurfactants were subjected to TLC (silica gel 60 F254) analysis using Chloroform: methanol: water (65:25:4, v/v) solvent system. The separated bands were visualized with iodine vapor. They were dissolved in chloroform: methanol (2:1, v/v) and subjected to FTIR analysis21 (Thermo Nicolet 6700) in the range of 4000–400cm−1. The extracted biosurfactants were also subjected to GCMS analysis for identification of component fatty acids (GCMS-Perkin Elmer Clarus 680) and the spectra were compared with the GCMS NIST (2008) library compounds.
Biodegradation of crude oil and determination of total petroleum hydrocarbon (TPH)To ascertain the crude oil degradation capacity of PA, PM and BM, the TPH content of residual crude oil was determined after culturing the bacteria in MSM with 1% crude oil for 13 days while the uninoculated medium served as control. 500mg of residual crude oil was taken from the medium and extracted consecutively with hexane, benzene, chloroform and methanol.22 The extracts were evaporated, pooled together and dissolved in n-pentane and separated into soluble and insoluble fractions and the TPH content was determined by the gravitational method as further (asphaltene). The TPH fraction was loaded on to a silica gel column (100–200mesh) to separate aliphatic and aromatic fractions by using hexane and benzene respectively. The aliphatic and aromatic fractions of TPH were analyzed by GCMS (PerkinElmer Clarus 680) and the spectra was compared with the library compounds (GC-MS NIST 2008). The percentage of crude oil degradation was calculated as [(control-treated)/control]*100.22 Quantification of hydroxylated aromatic compounds as an indicator of degradation of polycyclic aromatic hydrocarbon (PAH) was determined as resorcinol equivalent (RE)22 where Resorcinol served as a standard.
Identification of crude oil degrading bacteria by 16S rRNA gene sequencingGenomic DNA from the strains PA, PM and BM was isolated by the phenol chloroform extraction method.23 Amplification of 16S rRNA gene from genomic DNA was performed using universal forward and reverse primers fD1 (5′-GAGTTTGATCCTGGCTCA-3′) and rP2 (5′-ACGGCTACCTTGTTACGACTT-3′) in DNA thermo cycler (Eppendorf, Master cycler gradient). The amplified PCR products were purified and sequencing24 was carried out by Eurofins Genomics India Pvt Ltd, Bangalore. The nucleotide sequences obtained were compared with sequences of GenBank using nBLAST search. The phylogenetic tree was constructed by using MEGA 5 software version 5.1.25
ResultsIn this study, 12 bacterial strains were isolated from crude oil contaminated soil samples by using MSM with 1% crude oil. From these 12 strains, 3 potent strains with the isolate designation of PA, PM and BM were selected based on their improved crude oil degrading capacity. The capacity of these strains to use crude oil as sole carbon source was determined by analyzing the activity of catechol dioxygenase enzymes (C12O and C23O), characterizing the biosurfactants produced by them and by measuring the residual TPH content. The three strains were also subjected to molecular identification by 16S rRNA gene sequence analysis.
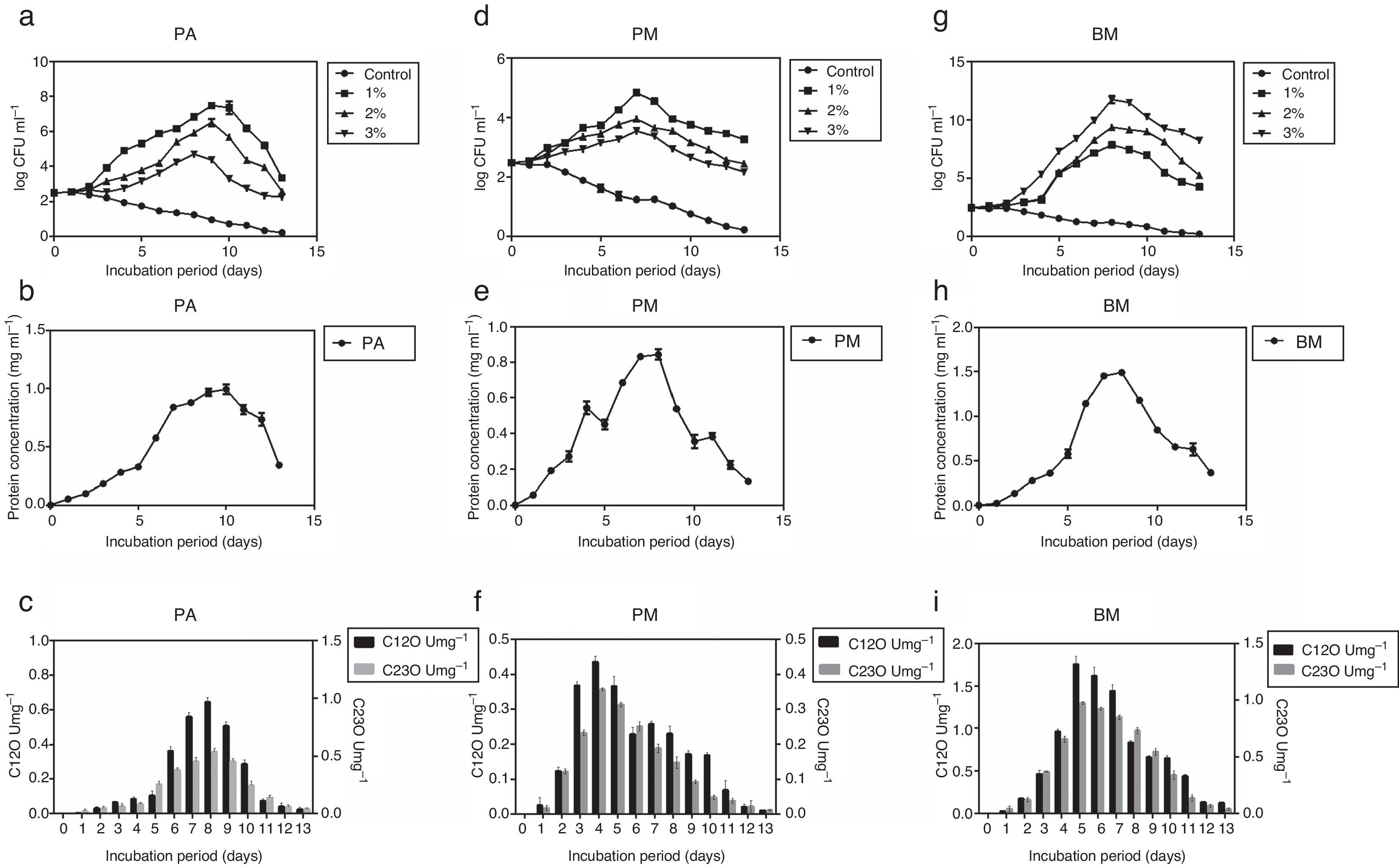
Determination of growth curve and C12O and C23O activityPure cultures of PA, PM and BM were allowed to grow on MSM containing different concentrations of crude oil (1, 2 and 3% (v/v) for 13 days and during this period growth rate of bacterial strains, their protein content and activity of C12O and C23O were determined at 24h intervals. Among the three strains, PA and PM showed maximum growth in media containing 1% crude oil while BM displayed maximum growth in media containing 3% crude oil. Exponential growth was observed between days 2 and 10 for PA, 2 and 7 for PM and 2 and 8 for BM (Fig. 1a, d and g). As PA and PM showed optimum growth in presence of 1% crude oil and BM with 3% oil, they were cultured in MSM with appropriate concentration of crude oil in order to determine the activity of enzymes –C12O and C23O. Concomitant increase in protein content (Fig. 1b, e and h) was also observed in all the three strains after the 5th day of incubation and peak protein levels were observed on days 10, 7 and 8 respectively for PA (0.991mgmL−1), PM (0.891mgmL−1) and BM (1.49mgmL−1). This trend clearly pointed to the ability of three bacterial strains to utilize crude oil as a source of carbon and energy. As could be seen in Fig. 1c, f and i, activity of C12O as well as C23O increased with days of incubation and the maximum activity was attained on day 8 for PA, day 4 for PM and day 5 for BM. This increasing trend in enzyme activity could be correlated well with the exponential growth shown by the three bacterial strains by utilizing crude oil as carbon source (Fig. 1a, d and g). Another interesting observation made in this study was the specific activity of C12O was higher than that of C23O in all the three strains (Fig. 1c, f and i). This increased activity of C12O signify the role of ortho- cleavage pathway as the predominant pathway of oil degradation. Among the three strains used in this study, BM exhibited increased activity of both C12O and C23O with maximum specific activity of 1.75U/mg−1 and 0.973U/mg−1 respectively. This finding may be correlated with the ability of BM to utilize higher concentration of crude oil and show maximum growth in MSM containing 3% crude oil. The other two strains PA and PM also exhibited moderate levels of activity of C12O and C23O. The maximum specific activity of C12O was 0.967 and 0.436Umg−1 and that of C23O was 0.537 and 0.356Umg−1 respectively in PA and PM.
Growth curves of the three bacterial strains – (a) PA, (d) PM and (g) BM in the presence of different concentrations of crude oil (1%, 2% and 3% v/v), protein content (mgmL−1) in (b, e and h) and Specific activities (U=1μmol of the product/min/mg protein) of C12O (Umg−1) and C23O (Umg−1) of bacterial strains grown in MSM with crude oil (concentration in %) – (c, f and i) PA-1%, PM-1% and BM-3%.
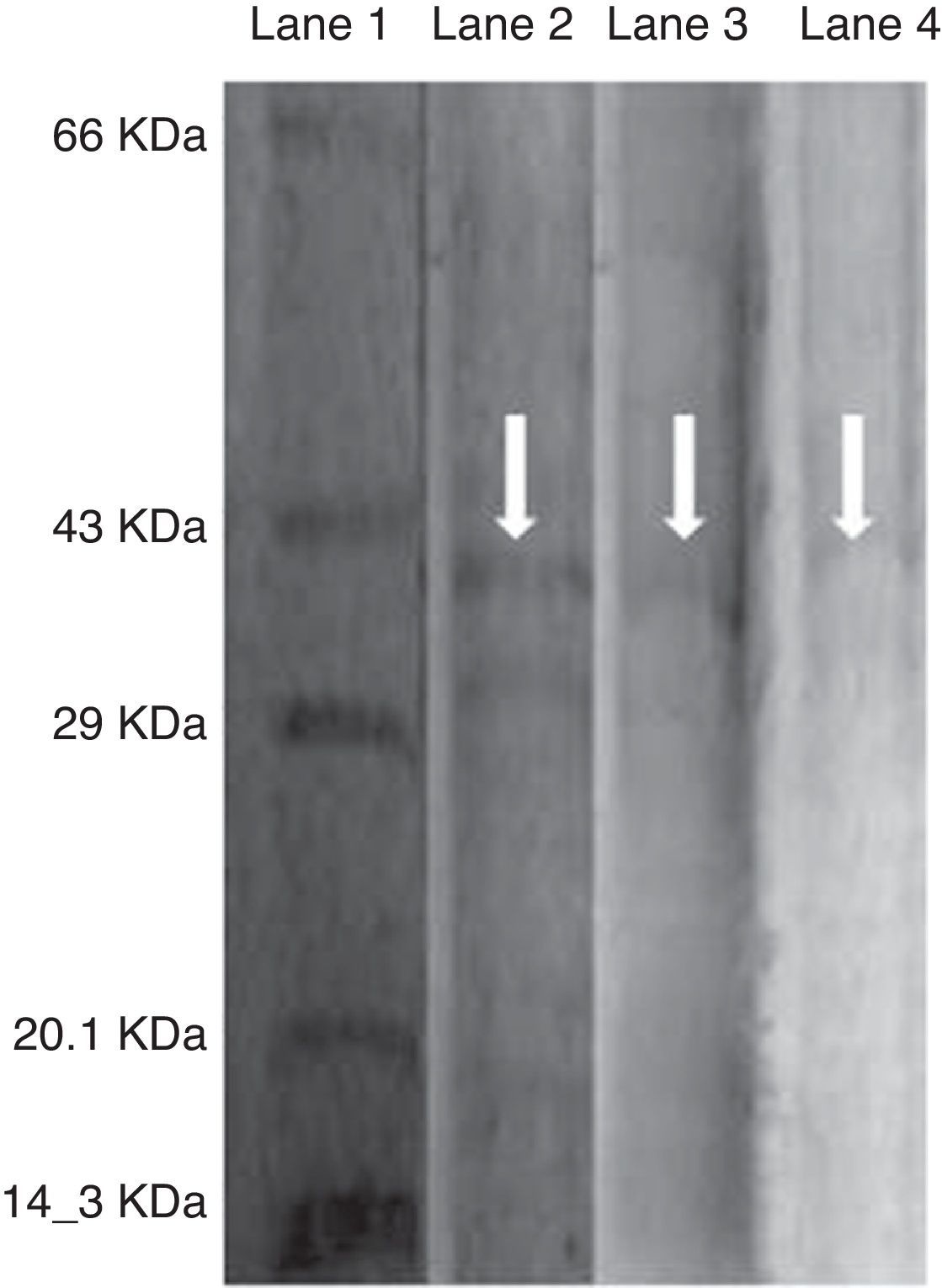
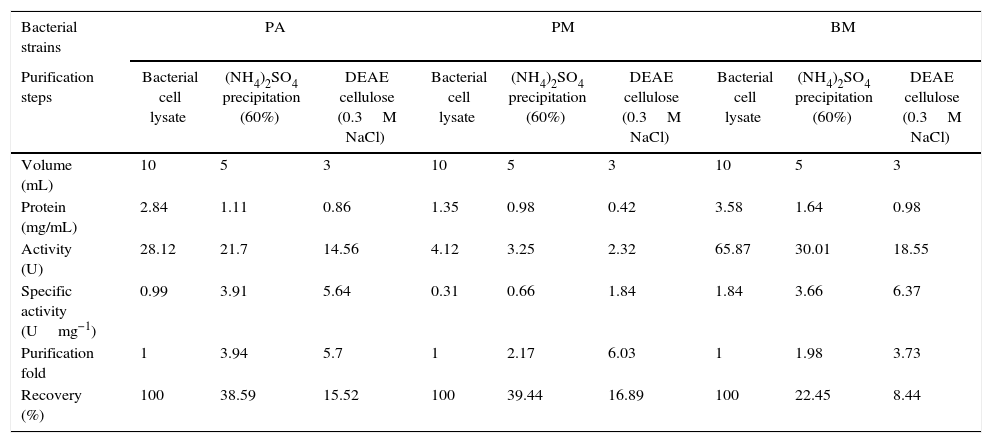
As the activity of C12O was found to be more than C23O in PA, PM and BM, partial purification of C12O alone was carried were subjected to ammonium suphate precipitation at 40%, 60% and 80% saturations and the precipitates obtained were checked for enzyme activity after desalting the pellets by dialysis. As the precipitate obtained at 60% saturation showed higher C12O activity, the same was used for further purification by ion-exchange chromatography using DEAE cellulose column. The column was eluted with 0.1–0.5M NaCl gradient and fractions obtained at 0.3M NaCl were found to exhibit better enzyme activity. The specific activity of C12O from PA, PM and BM in different purification steps are depicted in Table 1. Among the three strains, BM showed reasonably higher specific activity for C12O with 6.37Umg−1 followed by PA and PM with respective specific activities of 5.64 and 1.8Umg−1 following purification by ion exchange chromatography. The parially purified fraction of C12O from ion-exchange column showed a discrete protein band with an approximate molecular mass of 39 KDa in SDS-PAGE (Fig. 2).
Purification of C12O enzyme from PA (Corynebacterium aurimucosum), PM (Acinetobacter baumannii) and BM (Microbacterium hydrocarbonoxydans).
| Bacterial strains | PA | PM | BM | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Purification steps | Bacterial cell lysate | (NH4)2SO4 precipitation (60%) | DEAE cellulose (0.3M NaCl) | Bacterial cell lysate | (NH4)2SO4 precipitation (60%) | DEAE cellulose (0.3M NaCl) | Bacterial cell lysate | (NH4)2SO4 precipitation (60%) | DEAE cellulose (0.3M NaCl) |
| Volume (mL) | 10 | 5 | 3 | 10 | 5 | 3 | 10 | 5 | 3 |
| Protein (mg/mL) | 2.84 | 1.11 | 0.86 | 1.35 | 0.98 | 0.42 | 3.58 | 1.64 | 0.98 |
| Activity (U) | 28.12 | 21.7 | 14.56 | 4.12 | 3.25 | 2.32 | 65.87 | 30.01 | 18.55 |
| Specific activity (Umg−1) | 0.99 | 3.91 | 5.64 | 0.31 | 0.66 | 1.84 | 1.84 | 3.66 | 6.37 |
| Purification fold | 1 | 3.94 | 5.7 | 1 | 2.17 | 6.03 | 1 | 1.98 | 3.73 |
| Recovery (%) | 100 | 38.59 | 15.52 | 100 | 39.44 | 16.89 | 100 | 22.45 | 8.44 |
Several micro organisms that utilize petroleum hydrocarbons as their carbon source are known to produce biosurfactants of glycolipid or lipoprotein nature. In this study, biosurfactants produced by PA, PM and BM were characterized by physicochemical, biochemical, FTIR and GCMS analysis. The physicochemical properties of the biosurfactants were analyzed by determining the emulsification index (E24) % and oil displacement capacity of the culture supernatants of PA, PM and BM. The emulsification index for PA, PM and BM was 57, 45 and 69% respectively with BM showing higher index. The three bacterial strains also exhibited well pronounced oil displacement property with respective oil clearance zone of 16, 14 and 17mm. The chemical natures of the biosurfactants from culture supernatants of PA, PM and BM were identified qualitatively as glycolipids while they gave negative result for proteins. Further, quantitative analysis for glucose content in the culture supernatants of PA, PM and BM showed 135, 120 and 187mg/L of glucose respectively.
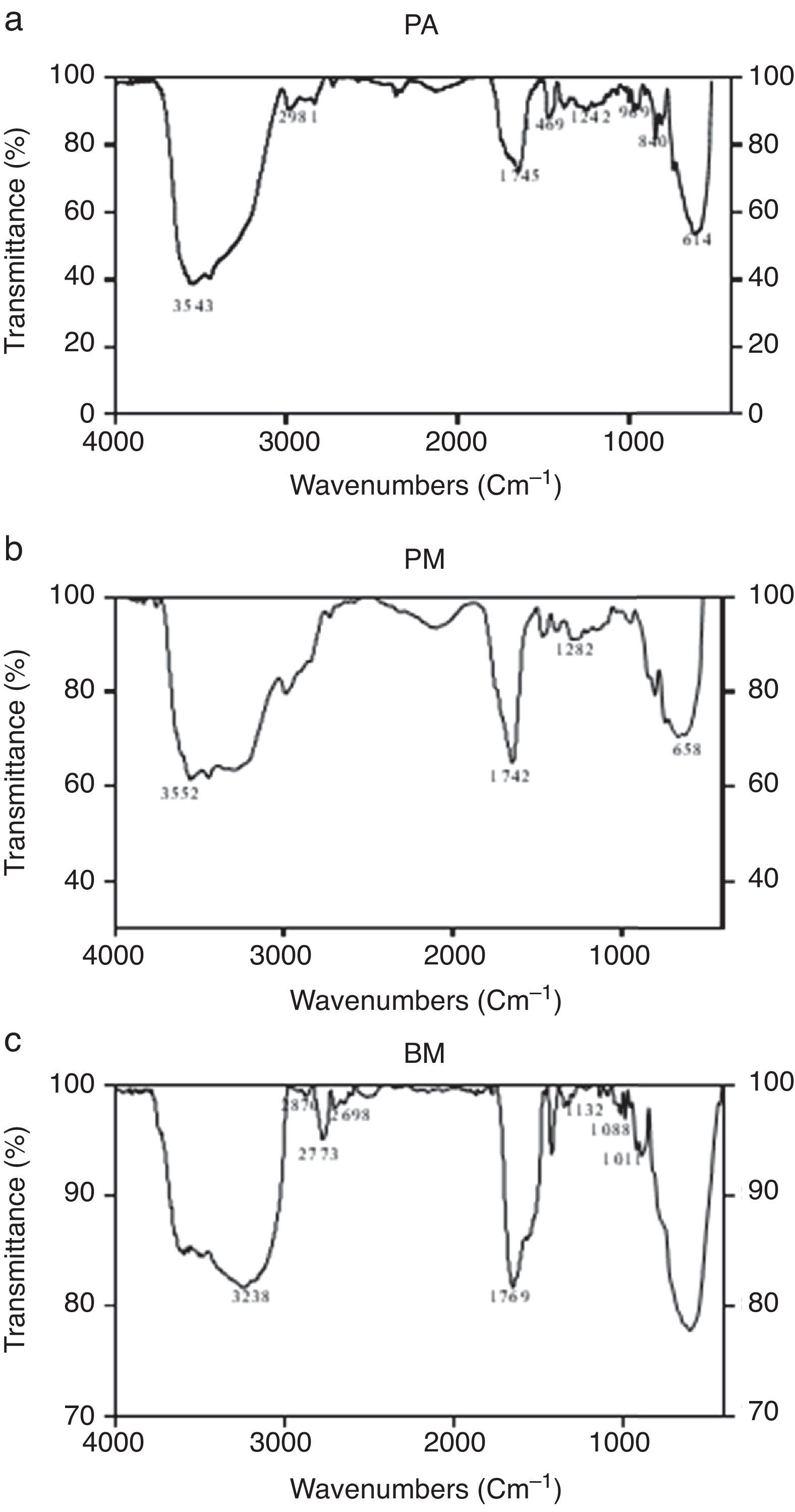
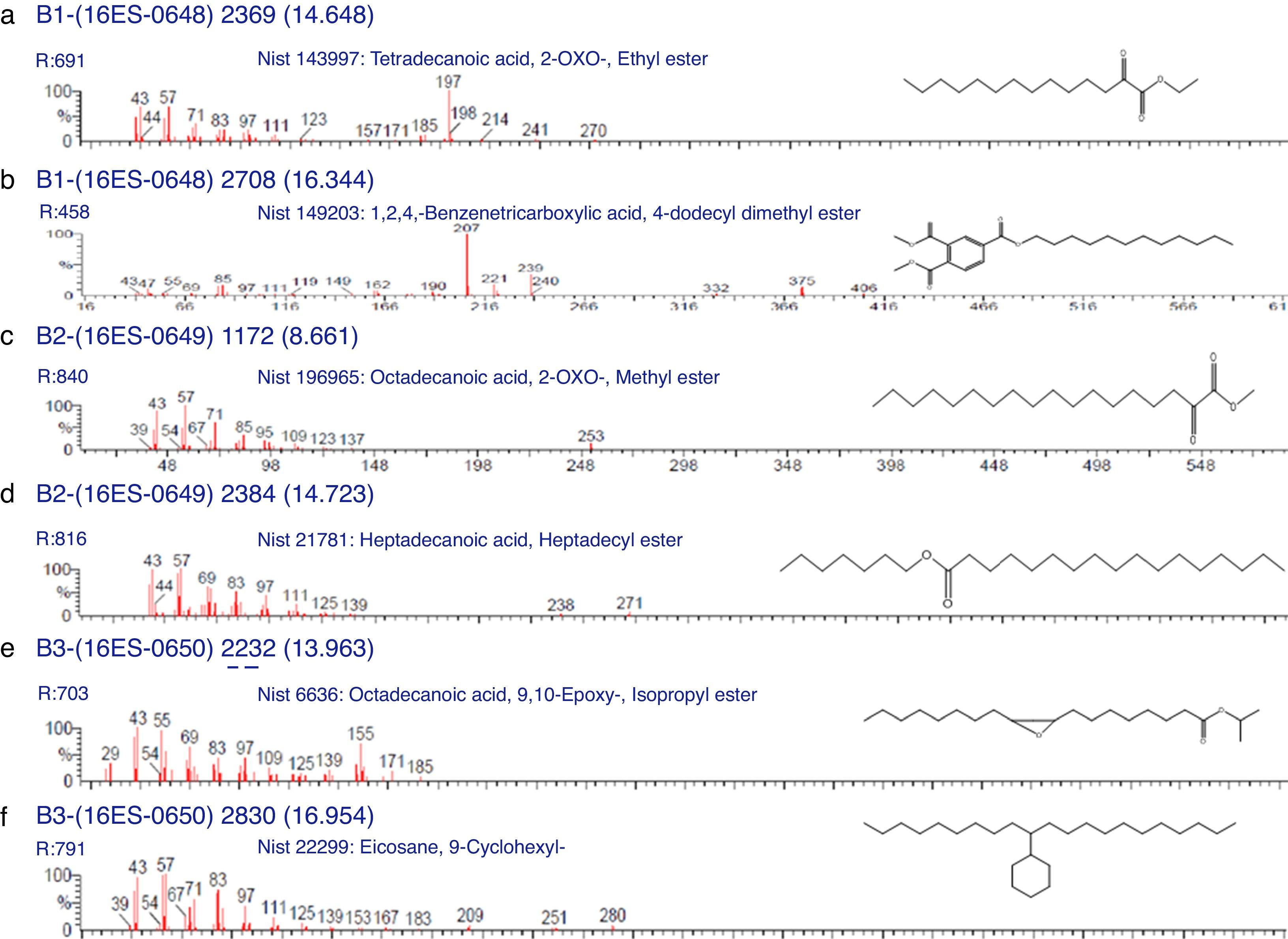
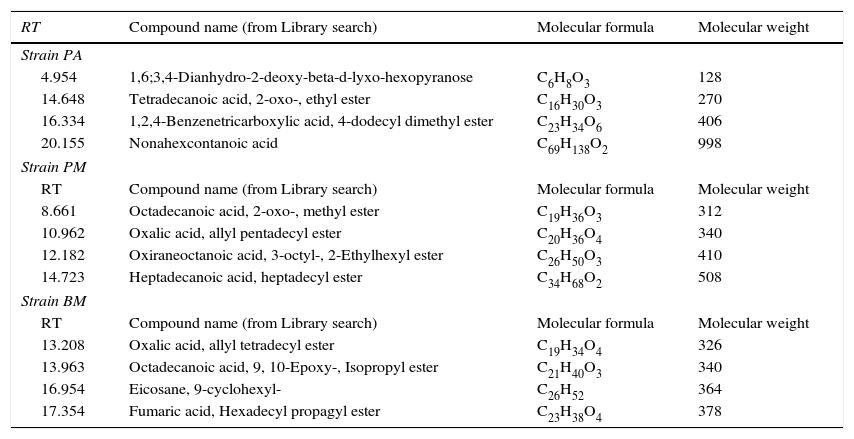
Characterization of biosurfactants by FTIR and GCMS analysisThe biosurfactants extracted from PA, PM and BM was subjected to FTIR analysis and the FTIR spectra are presented in Fig. 3. The broad band at 3543, 3552 and 3238cm−1 observed respectively for PA BM and PM should be assigned to the O–H stretching vibration in the chemical structures of the biosurfactants. The strong peaks observed at 2981, 1469cm−1 for PA, 2960cm−1 for PM and 2870, 2773, 2698cm−1 from BM correspond to the C–H stretching vibrations of the CH2 and CH3 hydrocarbon chains. The characteristic peak displayed at 1745, 1742 and 1769cm−1 respectively for PA, PM and BM relates to the CO stretching vibrations of the carbonyl groups. Same way the peaks observed at 1242 for PA, 1282 for PM and 1132, 1088 and 1011cm−1 for BM represent C-O stretching bands that confirm the presence of glycosidic bonds formed between carbon atoms and hydroxyl groups. The biosurfactants that were extracted from PA, PM and BM was further analyzed by GCMS analysis to detect the presence of fatty acids and the data are presented in Fig. 4. The GCMS spectra of PA PM and BM showed four major peaks and the corresponding fatty acids/fatty acid esters are listed in Table 2.
List of compounds identified in Biosurfactants from PA (Corynebacterium aurimucosum), PM (Acinetobacter baumannii) and BM (Microbacterium hydrocarbonoxydans).
| RT | Compound name (from Library search) | Molecular formula | Molecular weight |
|---|---|---|---|
| Strain PA | |||
| 4.954 | 1,6;3,4-Dianhydro-2-deoxy-beta-d-lyxo-hexopyranose | C6H8O3 | 128 |
| 14.648 | Tetradecanoic acid, 2-oxo-, ethyl ester | C16H30O3 | 270 |
| 16.334 | 1,2,4-Benzenetricarboxylic acid, 4-dodecyl dimethyl ester | C23H34O6 | 406 |
| 20.155 | Nonahexcontanoic acid | C69H138O2 | 998 |
| Strain PM | |||
| RT | Compound name (from Library search) | Molecular formula | Molecular weight |
| 8.661 | Octadecanoic acid, 2-oxo-, methyl ester | C19H36O3 | 312 |
| 10.962 | Oxalic acid, allyl pentadecyl ester | C20H36O4 | 340 |
| 12.182 | Oxiraneoctanoic acid, 3-octyl-, 2-Ethylhexyl ester | C26H50O3 | 410 |
| 14.723 | Heptadecanoic acid, heptadecyl ester | C34H68O2 | 508 |
| Strain BM | |||
| RT | Compound name (from Library search) | Molecular formula | Molecular weight |
| 13.208 | Oxalic acid, allyl tetradecyl ester | C19H34O4 | 326 |
| 13.963 | Octadecanoic acid, 9, 10-Epoxy-, Isopropyl ester | C21H40O3 | 340 |
| 16.954 | Eicosane, 9-cyclohexyl- | C26H52 | 364 |
| 17.354 | Fumaric acid, Hexadecyl propagyl ester | C23H38O4 | 378 |
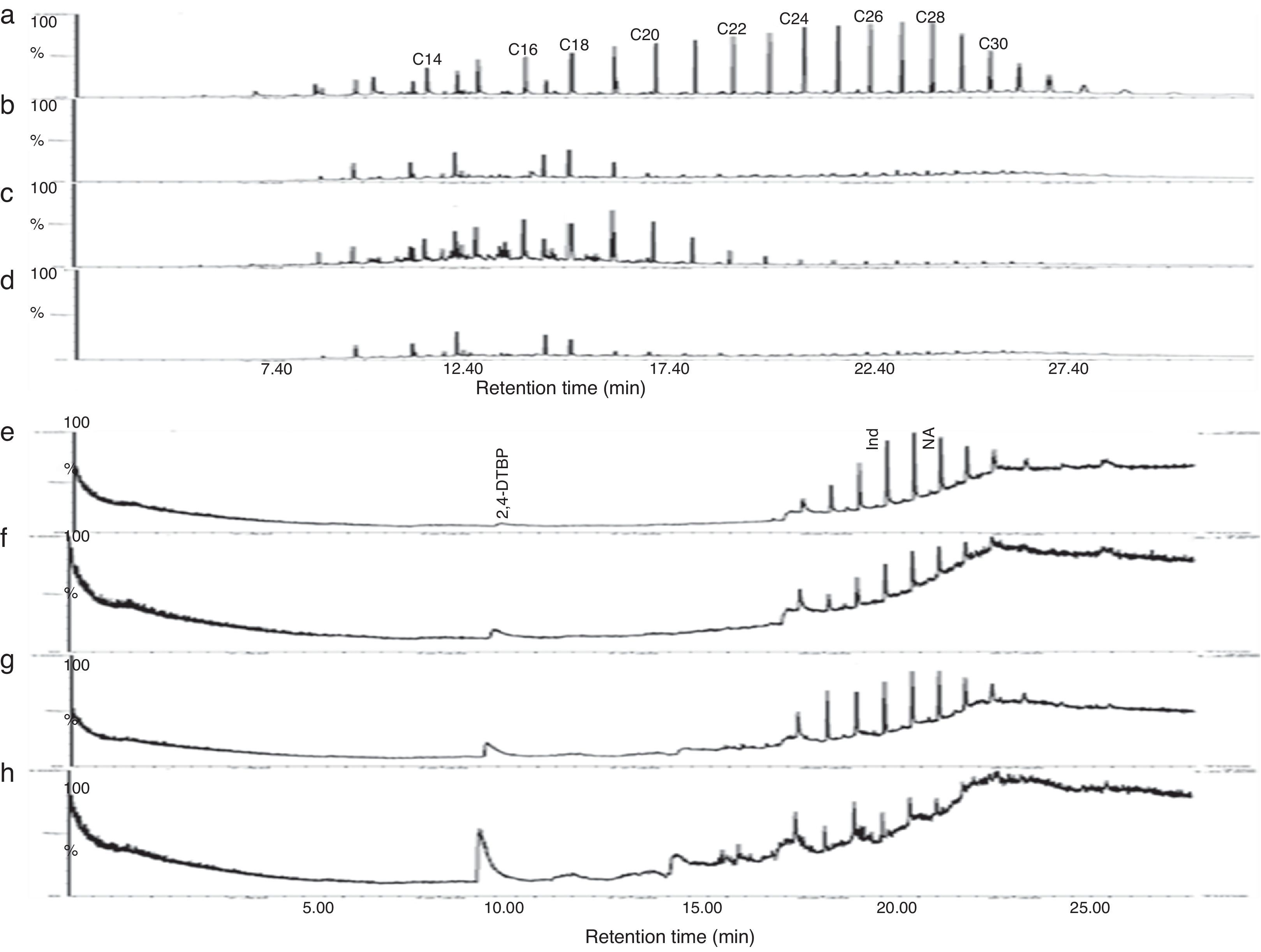
Gravitational analysis of TPH from un-inoculated crude oil indicated the presence of 0.282g of aliphatic, 0.098g of aromatic and 0.04g of NSO compounds along with 0.059g of asphaltenes. On the other hand, drastic reduction in the TPH content was observed at the end of 13 days incubation with PA, PM and BM. The aliphatic, aromatic, NSO and asphaltene fractions of residual TPH in presence of BM was found respectively as 0.042, 0.014, 0.024, 0.036g and 0.048, 0.053g, 0.063g and 0.044g in presence of PA while with PM it was 0.114, 0.073, 0.027 and 0.035g. From these observations it could be assumed that BM was more competent in utilizing crude oil and it brought about 74% total degradation as compared to un-inoculated control while PA and PM brought about 52 and 43% degradation respectively. Further the GC chromatogram of aliphatic and aromatic fraction of TPH from un-inoculated control and flasks inoculated with PA, PM and BM is presented in Fig. 5. It was observed that BM brought about substantial degradation of aliphatic compounds with 81% of C14, 12% of C16, 27% of C18 and more than 85% of C20–C30 (Fig. 5d) as compared to un-inoculated control (Fig. 5a). Same way, PA also showed enhanced degradation with 87% of C14, 34% of C16, 68% of C18 and more than 85% of C20–C30 (Fig. 5b). Whereas PM brought about lesser effect with 3.5% degradation of C14, 11% of C16, 8% of C18, 64% of C20, 81% of C22, and 85% of C24–C30 (Fig. 5c). Similar effects were observed with the percentage degradation of aromatic compounds-indene and naphthalene in presence of BM which was 90 and 87% respectively (Fig. 5h) as compared to un-inoculated control (Fig. 5e) and in the presence of PA it was of 42 and 52% (Fig. 5f) while with PM it was 11 and 31% respectively (Fig. 5g). Also, an increase in the formation of intermediate metabolite 2, 4-di-tert-butyl phenol (2,4-DTBP) during the degradation of naphthalene was observed more prominently in presence of BM (Fig. 5h). The intermediates formed during the degradation PAH were determined as resorcinol equivalent (RE) which was found to be higher with BM (87.43μgmL−1) followed by PA (67.68μgmL−1) and PM (65.89μgmL−1) as compared to un-inoculated control (1.76μgmL−1).
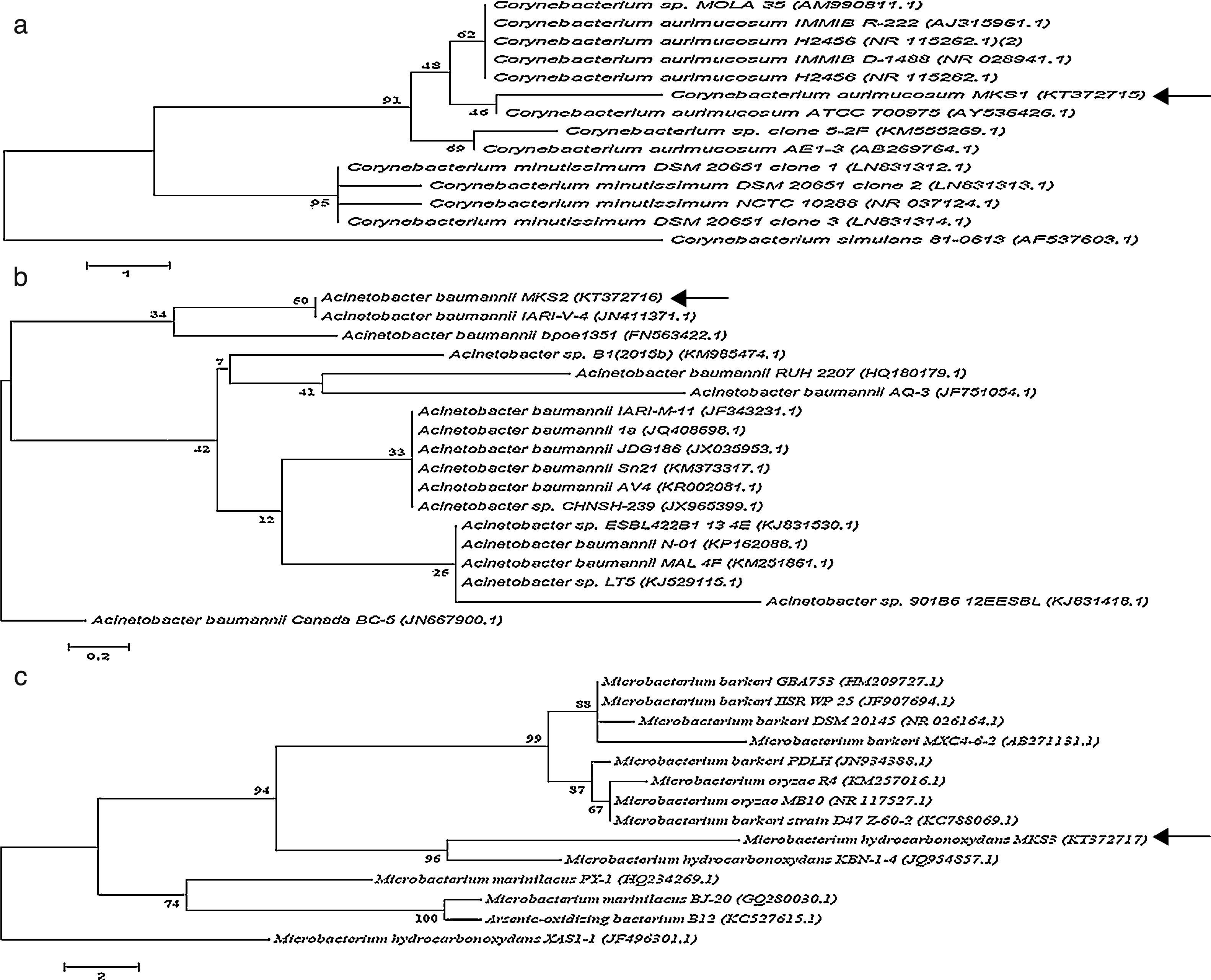
Identification of bacteriaThe three bacterial isolates-PA, PM and BM were subjected to morphological and molecular identification. Morphological analysis indicated that all the three strains were aerobic, non-pigmented and non-sporulating bacteria. By Gram's staining procedure, PA and BM were found to be gram-positive rods while PM was gram-negative rod. Further, molecular characterization of PA, PM and BM was performed by 16S rRNA gene sequence analysis and the nucleotide sequences (958bp, 1028bp and 803bp) of the PA, PM and BM obtained were deposited in the National center for Biotechnology information (NCBI Gen Bank) under the respective accession numbers KT372715, KT372716 and KT372717 and designated as Corynebacterium aurimucosum MKS1, Acinetobacter baumannii MKS2 and Microbacterium hydrocarbonoxydans MKS3 respectively. Phylogenetic trees of PA, PM and BM was constructed and shown in Fig. 6. The 16S rRNA gene sequence of PA revealed 97% identity to C. aurimucosum ATCC 700975 (AY536426.1), PM showed 99% identity to A. baumannii IARI-V-4 (JN411371.1) and BM showed 99% identity to M. hydrocarbonoxydans KBN-1-4 (JQ954857.1).
Phylogenetic position of bacterial strains PA, PM and BM and representatives of some other related taxa based on the 16S rRNA gene sequences. Corynebacterium simulans 81-0613 (AF537603.1), Acinetobacter baumannii Canada BC-5 (JN667900.1) and Microbacterium hydrocarbonoxydans XAS-1 (JF751054.1) served as external reference for PA, PM and BM respectively. Bar (1, 0.2 and 2 for PA, PM and BM respectively) substitution per nucleotide position. Bootstrap values expressed as percentages of 1000 replications.
Role of microbial activity in the biodegradation of hydrocarbons has been well recognized for more than a century.1 The native bacterial populations in the crude oil contaminated soil sites possess the capacity to mineralize hydrocarbons.26 In the present study, the growth curves of PA, PM and BM depicted that the cells adapted well to the media containing crude oil within 2 days and exhibited exponential growth between 7 and 10 days. It was interesting to observe that BM displayed maximum growth in media supplemented with 3% crude oil suggesting its superior ability to utilize higher concentration of crude oil while PA and PM showed maximum growth in presence of 1% crude oil. Similar observations have been made with Bacillus sp. S6 and S35 which showed exponential growth in MSM containing 1% crude oil up to day 7.27 In yet another study, Pseudomonas sp. BP10 and Rhodococcus sp. NJ2 exhibited slow growth initially between 0 and 20 days in MSM containing 2% crude oil and showed exponential growth between 20 and 25 days.22 The concomitant increase in the protein content of PA, PM and BM observed in this study correlated well with increasing cell count and their ability to utilize crude oil as a source of carbon and energy. Also, moderate activity enzymes C12O as well as C23O enzymes was observed in all the three strains which correlated well with the exponential growth shown by PA, PM and BM utilizing crude oil. Another interesting observation made in this study was that the specific activity of C12O was higher than that of C23O in all the three bacterial strains. This increased activity of C12O signify the role of ortho-cleavage pathway as the predominant pathway of oil degradation. Among the three strains used in this study, BM exhibited higher activity of C12O as well as C23O which validated the superior capacity of BM to show maximum growth in MSM with 3% crude oil. Similar to our findings, increased specific activity for C12O than C23O was exhibited by Rhodococcus pyridinivorans BP101 and Pseudomonas sp. NJ2 strains cultured on MSM with crude oil.22 In a different study, increased activity of C12O (1.03μmolmin−1mg−1) than C23O (0.37μmolmin−1mg−1) was observed in Stenotrophomonas maltophilia KB2 cultured on MSM containing phenol as carbon source.28
Partial purification of C12O from PA, PM and BM in this study by using ammonium sulphate precipitation followed by ion-exchange chromatography specified that the activity of the enzyme was prominently higher in bacterial strain BM followed by PA. This increased activity of C12O can be correlated with the increased growth rates shown by BM in 3% crude oil containing MSM in this study. Our findings on increasing specific activity of C12O at different purification steps draw a parallel to the observations made in a recent study where the specific activity of C12O from crude cell lysate of Rhodococcus sp NCIM 2891 increased from 0.78 to 1.83Umg−1 after ion exchange chromatography.14 Similarly, the activity of crude C12O from Acinetobacter radioresistens was shown to increase from 1.63 to 6.3 and then to 24.5Umg−1 respectively after purification by ion exchange and gel permeation chromatography.29 The molecular mass of parially purified fraction of C12O enzyme from all the three strains PA, PM and BM in this study was found to be ∼39KDa and this corroborate well with the observations made on C12O from A. radioresistens29 and Rhodococcus rhodochrous.30
Several micro organisms including Pseudomonas sp., Rhodococcus sp. that utilizes petroleum hydrocarbons as their carbon source are known to produce biosurfactants of glycolipids or lipoprotein nature.22 In the present study, the biosurfactants of from PA, PM and BM were qualitatively identified to be glycolipid in nature. The biosurfactants also exhibited substantial emulsification index as well as oil displacement ability. The surface tension attained by the biosurfactants displaces the oil and the diameter of the clearance zone on the oil surface gives an indirect indication of biosurfactant activity.17 Among the three strains used in this study, BM displayed superior biosurfactant activity (E24=69%) and oil displacement capacity (17mm). This enhanced surfactant production can be correlated with the superior growth rate of BM observed in this study by utilizing higher concentration of crude oil (3%). Similar observations have been made with different bacterial species such as Streptomyces matensis PLS-131 and P. aeruginosa32 which produced biosurfactants of glycolipid nature. In a different study, the crude oil degrading capacity of Acinetobacter calcoaceticus was correlated with the level of biosurfactants produced (E24= 64%) and superior oil displacement activity (7.2cm).33 FTIR has been widely used to characterize the surface groups as the IR transmission spectra presents peaks characteristic of specific chemical bonds.34 In the present study the FTIR spectra of extracted biosurfactants from PA, PM and BM indicated the presence of glycosidic bonds and thereby confirming the glycolipid nature of the biosurfactants. Similar to our observations earlier workers have confirmed the glycolipid nature of biosurfactants produced by Bacillus megaterium7 by FTIR analysis. Subsequent GCMS analysis of the extracted biosurfactants from PA, PM and BM confirmed the presence of fatty acids/fatty acid esters. The presence of decanoic acid esters in the biosurfactant preparations observed in this study can be correlated with the predominance of methyl esters of decanoic acids observed in the biosurfactants from A. Niger, A. flavus35 and Bacillus sp.36
The residual TPH content following crude oil degradation by PA, PM and BM was analyzed using gravitational method which indicated that BM utilized TPH more effectively than PA and PM. Similar observations have been made in previous studies with various other bacterial strains. It was shown that P. aeruginosa DQ8 degraded 79.3% of TPH in MSM with 10% of crude oil.37 Similarly, A. calcoaceticus BS and Alcanivorax dieselolei PG-12 were shown to degrade 82% and 71% of 1% (v/v) crude oil respectively owing to their high emulsification activity and biosurfactant production.38 In this study, it was observed that the degradation of aliphatic compounds of TPH was effectively brought about by both PA and BM (more than 80% of C14 and 85% of C20-C30 compounds) more than PM. Similar observations have been made in previous studies where Rhodococcus sp. Moj-3449 degraded compounds in the range of C14-C1939 while A. calcoaceticus and Alcaligenes odorans preferred C17-C24 compounds of crude oil.38 Same way, the degradation of aromatic compounds of TPH by PA, PM and BM when analyzed, it was observed that BM was more competent in degrading poly aromatic hydrocarbons (more than 85%) as compared to PA and PM which could degrade only to the extent of 40 and 10% respectively. In an earlier report, Streptomyces spp. was shown to degrade poly aromatic hydrocarbon naphthalene to produce 2, 4-di-tert-butylphenol (2,4-DTBP) as byproduct.40 As the crude oil is a mixture of hydrocarbons, it is difficult to identify the intermediates formed during the degradation of crude oil. Since the aromatic compounds are more unmanageable than alkanes, the activity of dioxygenase41 and the production of hydroxylated aromatic compounds were used as a tool to trace the initial pathways of oil degradation. In this study, BM produced more resorcinol equivalent intermediates than PA and PM during the 13 days incubation period which suggested that BM possesses higher crude oil degrading potential than PA and PM. Similar to our findings, considerable increase in resorcinol equivalent intermediates were observed with Pseudomonas sp. (86.7μgmL−1) and Rhodococcus sp. (82.5μgmL−1) following 25 days of incubation with crude oil.22
Based on the molecular characterization by 16S rRNA gene sequence analysis, the three bacterial strains – PA, PM and BM were identified respectively as C. aurimucosum MKS1 (KT372715), A. baumannii MKS2 (KT372716) and M. hydrocarbonoxydans MKS3 (KT372717).
ConclusionThis study focused on three potent oil degrading bacteria strains PA, PM and BM that were isolated from oil polluted soil. These strains displayed substantial ability to utilize crude oil as their carbon source and among the three, BM exhibited superior capacity to degrade crude oil. As the dioxygenase enzymes C12O and C23O are known to play a vital role in the biodegradation of aromatic molecules, increased activity of C12O than C23O that was observed in this study in PA, PM and BM specify that the ortho-cleavage pathway as the predominant pathway of oil degradation by these bacteria. Also, their ability to produce biosurfactants seems to compliment their biodegradation potential. The residual TPH content analysis and GCMS analysis also confirmed the efficiency of crude oil degradation by the bacterial strains. The bacterial strains PA, PM and BM were identified as C. aurimucosum MKS1, A. baumannii MKS2 and M. hydrocarbonoxydans MKS3 respectively by the 16S rRNA gene sequence analysis. Thus, results from this study convincingly specify that such bacteria with potent oil degrading capacity may be vibrantly used for clearing unforeseen oil spillages.
Conflicts of interestThe authors declare no conflicts of interest
The authors duly acknowledge instrument support from the Department of Biochemistry and Molecular Biology, Pondicherry University, India and DST-FIST, Government of India. The first author (Muthukamalam S) conveys special thanks to UGC, New Delhi, India, for financial support in the form of Junior Research Fellowship (CSIR-UGC-JRF 316979; Ref no. 21/12/2014(ii)EU-V). The authors convey special thanks to Indian Oil Corporation Limited (IOCL), Chennai, Tamil Nadu, India for providing soil samples and crude oil.