Fecal bacteria are considered to be a potential reservoir of antimicrobial resistance genes in the aquatic environment and could horizontally transfer these genes to autochthonous bacteria when carried on transferable and/or mobile genetic elements. Such circulation of resistance genes constitutes a latent public health hazard. The aim of this study was to characterize the variable region of the class 1 integron and relate its genetic content to resistance patterns observed in antimicrobial-resistant Escherichia coli isolated from the surface waters of Patos Lagoon, Southern Brazil. Genetic diversity of the isolates and presence of the qacEΔ1 gene, which confers resistance to quaternary ammonium compounds, were also investigated. A total of 27 isolates were analyzed. The variable region harbored dfrA17, dfrA1 and dfrA12 genes, which confer resistance to trimethoprim, and aadA1, aadA5 and aadA22 genes that encode resistance to streptomycin/spectinomycin. Most of the isolates were considered resistant to quaternary ammonium compounds and all of them carried the qacEΔ1 gene at the 3′ conserved segment of the integron. ERIC-PCR analyses of E. coli isolates that presented the integrons showed great genetic diversity, indicating diverse sources of contamination in this environment. These results suggest that fecal bacteria with class 1 integrons in aquatic environments are potentially important reservoirs of antibiotic-resistance genes and may transfer these elements to other bacteria that are capable of infecting humans.
The rapid spread of antimicrobial resistance in bacteria is an ever-increasing global concern. Further, antimicrobial resistance is no longer restricted to medical or veterinary settings and has also been reported in many aquatic environments such as rivers,1 surface waters,2 sewage treatment plants3 and hospital effluents.4–6
Antimicrobial resistance genes are mainly carried on mobile genetic elements such as plasmids and transposons. Integrons are natural recombination systems that are able to capture, mobilize and express several gene cassettes encoding antimicrobial resistance, and are often found inserted into these mobile elements.7 Three classes of integrons are responsible for multi-drug resistance and are classified based on the integrase gene sequence they carry. Class 1 integrons are widely found in Gram-negative bacteria and are associated with both an increase in and spread of antimicrobial multi-drug resistance throughout the world.8
Most integrons are of the class 1 type and harbor two conserved segment (CS) regions, one each in the 5′ and 3′ regions. The 5′-conserved segment (5′-CS) contains an intI gene that encodes an integrase (intI1), a recombination site (attI) and a common promoter that is responsible for the expression of the captured gene cassettes, whereas the 3′-conserved segment (3′-CS) includes a gene conferring resistance to quaternary ammonium compounds (qacEΔ1), a sulfonamide resistance gene (sul1) and an ORF (orf5) of unknown function.9,10 These segments are separated by a variable region, wherein one or more gene cassettes are inserted.
Investigations into antimicrobial resistance in aquatic environments have mainly focused on bacteria of fecal origin as they are used as indicators of pollution, may be associated with infectious diseases and are also capable of transferring resistance at the host–environment interface.11,12Escherichia coli is a member of the natural gastrointestinal microbiota and is also a key indicator of fecal contamination in water bodies. Resistant E. coli may enter aquatic environments through the discharge of treated effluent from wastewater treatment plants,3 field runoffs, and from untreated effluents.13 It was recently found that some persistent strains of E. coli can survive for between 4 and 14 days in aquatic environments with low levels of fecal bacteria,14 and can thus, play an important role in the dissemination of antimicrobial resistance in this environment.
Studies that investigate antimicrobial resistance in natural ecosystems are of great importance in identifying environmental reservoirs of resistance and also contribute toward understanding the routes of dissemination of these resistant bacteria. Therefore, in order to develop a better understanding of the spread of antimicrobial resistance genes in surface waters, this study aimed to characterize the variable region of the class 1 integron and relate the gene cassettes found therein to the antimicrobial susceptibility pattern of resistant E. coli isolated from the surface water of the Patos Lagoon in Southern Brazil. The presence of the qacEΔ1 gene and susceptibility to quaternary ammonium compounds were also assessed.
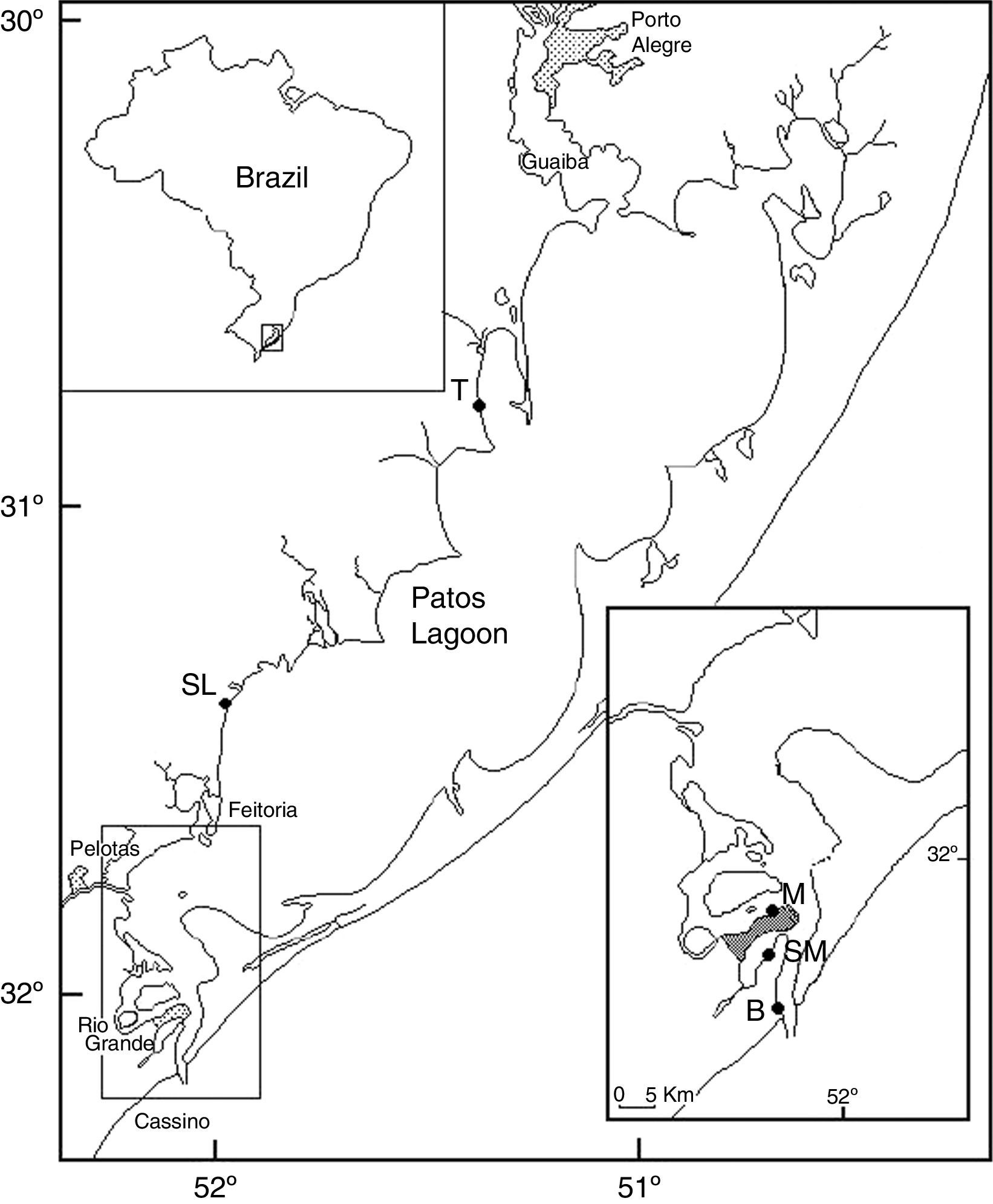
Materials and methodsIsolation and characterization of E. coli strainsIn a previous study, 441 E. coli were isolated from surface water at five sampling sites on the Patos Lagoon in Southern Brazil (Fig. 1).15 The Patos Lagoon is located between parallels 30°30′S and 32°12′S, is a semi-closed microtidal coastal lagoon, and its water circulation is mainly controlled by the northeast–southwest wind regime and freshwater input tributaries. The lagoon surface area is around 10,230km2 and extends in a northeast–southwest direction from the delta of the Guaíba Lake to the Atlantic Ocean near Rio Grande.17 This water system receives a variety of urban, industrial, and agricultural inputs, including those from the petrochemical industry in the cities of Porto Alegre and Rio Grande. Uncontrolled settlements, the use of agricultural fertilizers and pesticides near its shores, discharge of domestic and industrial effluents into the estuarine zone of the lagoon, among other factors, have contributed to the degradation of this lagoon; and the resultant adverse effects have intensified in the recent decades. Organic and fecal-bacterial contamination and excessive concentration of chemical compounds such as ammonium and phosphate due to the discharge of untreated domestic sewage along the western shore of the lagoon have led to eutrophication; nevertheless, this area is also very popular for fishing and recreation.18
Sampling sites in Patos Lagoon, Brazil. Sampling site Barra (B), Saco da Mangueira (SM) and Museum (M) located at the estuary of Patos Lagoon, at Rio Grande city; city of São Lourenço do Sul (SL); city of Tapes (T) (adapted).16
Surface water samples were collected four times from each sampling site between June 2007 and March 2008. The samples (4L; at 0.3m depth) were collected in sterile plastic bottles and stored at 4°C in the dark until analysis. All samples were analyzed within 8h after collection. Each water sample was diluted in 0.1% peptone water (10−1, 10−2, 10−3, 10−4, 10−5, 10−6) and filtered through a 0.45μm mixed-ester filter; this filter was then placed onto m-Endo Les Agar (Himedia) and incubated at 44.5°C for 24h.19 Pink colonies with a metallic sheen were inoculated onto Eosin Methylene Blue Agar (EMB, Himedia) and incubated at 37°C for 24h. After isolation of characteristic colonies, presumptive E. coli colonies were cryopreserved in Brain Heart Infusion Broth (BHI, Himedia) containing 15% glycerol and stored at −20°C.
Isolates were identified using Gram staining and biochemical tests (indole test, methyl red test, Voges–Proskauer test, citrate test, triple sugar iron agar, motility test, urease test and growth on presence of d-sorbitol). Antimicrobial resistance patterns were determined by the agar diffusion method on Mueller-Hinton Agar (Oxoid, Basingstoke, UK) using the following 17 antimicrobial agents at concentrations indicated in parentheses: ampicillin (10μg), amoxicillin/clavulanic acid (20μg/10μg), piperacillin/tazobactam (100μg/10μg), aztreonam (30μg), imipenem (30μg), cefoxitin (30μg), ceftazidime (30μg), cefotaxime (30μg), cefepime (30μg), amikacin (30μg), gentamicin (30μg), chloramphenicol (30μg), tetracycline (30μg), trimethoprim/sulfamethoxazole (1.25μg/23.75μg), norfloxacin (30μg), sulfonamide (250μg), streptomycin (10μg) and spectinomycin (100μg).20 Isolates that showed resistance to at least three different classes of antimicrobials were considered multiresistant.1 The presence of the class 1 integron was tested by PCR using primers for the integrase (intI1) gene21 with PCR conditions were previously established.6
Of the isolates, 157 were resistant to at least one antimicrobial agent and were tested for the presence of intI1 gene. The intI1 gene was detected in 62 isolates, and 43 of these isolates that were resistant to two or more antimicrobials agents were selected for analyzing the variable region of the class 1 integron.
Evaluating genetic diversity of E. coli isolates using ERIC-PCR (Enterobacterial Repetitive Intergenic Consensus Sequence PCR)DNA was extracted as described previously (Misbah et al., 2005)22 and the variable region was amplified using primers ERIC1 and ERIC2.23 PCRs were performed in a volume of 25μL containing 1× reaction buffer, 6.5mM MgCl2, 125μM dNTPs (Biolabs), 400ng of each primer, 1U Taq DNA polymerase (Invitrogen Life Technologies) and 7μL of genomic DNA (approximately 100ng). The amplification conditions were as follows: initial denaturation at 94°C for 7min, followed by 35 cycles of denaturation for 1min at 94°C, annealing for 1min at 51°C, and extension for 3min at 72°C, with a final extension of 15min at 72°C. In all ERIC-PCRs E. coli (ATCC 25922) was used as a positive control and also for standardization and evaluation of reproducibility.
The amplified products were separated on a 2% agarose gel in Tris–borate buffer for 4h at 80V. The gels were stained with ethidium bromide (0.5μg/mL), visualized on a UV transilluminator and documented using the Kodak 1D image analysis software. The ERIC-PCR fingerprints were transformed onto a similarity matrix based on the presence or absence of each band. Only visible bands in the ERIC-PCR fingerprints were used to construct the similarity matrix and the dendrogram. Similarity among profiles was determined using the Jaccard coefficient and dendrograms were generated using the unweighted pair-group with mathematic average (UPGMA) method. All calculations were performed using the NTSYSPC version 2.0 software (Applied Biostatistics).
Amplification and sequencing of the variable region of the class 1 integron and amplification of the qacΔE1 geneThe variable region of the class 1 integron was amplified using previously established primers 5CS′/3CS′.9 The extraction of DNA was performed as previously described and PCRs were performed in a volume of 50μL containing 2μL genomic DNA (approximately 100ng), 1× reaction buffer, 1U of Taq DNA polymerase, 200μM of each dNTP, 2.5mM MgCl2 and 5μM of each primer. PCR conditions were as follows: initial denaturation of 5min at 94°C, followed by 35 cycles of 1min at 94°C, 1min at 50°C, and 2min at 72°C, with a final extension of 10min at 72°C. E. coli ATCC 25922 and Pseudomonas aeruginosa blaVIM-1 were used as negative and positive controls, respectively. The PCR products were analyzed on a 1% agarose gel containing 0.5μg/mL ethidium bromide, visualized with UV light and analyzed using image analysis Kodak 1D software (version 3.5.2). PCR product size was estimated using a 100bp Ladder (New England Biolabs).
The PCR products were purified using the Invisorb Fragment CleanUp kit (Stratec Molecular, Berlin). DNA sequencing was performed on both strands using primers used for amplification using an ABI Prism Big Dye Terminator Cycle Sequencing Kit and analyzed on an ABI Prism 310 Genetic Analyzer (Applied Biosystems). The sequences were analyzed using BioEdit and aligned with GenBank data using the nucleotide Basic Local Alignment Search Tool (BLAST) software. A sequence was tagged as that of a gene if the similarity with GenBank sequence was at least 98%. Nucleotide sequences were deposited in the GenBank database under accession numbers KJ569553, KJ569554, KJ569555, KJ569556, KJ569557, KJ569558, KJ569559, KJ569560, KJ569561, KJ547712, KJ547713, KJ547714, KJ547715, KJ547716, KJ547717, KJ547718, KJ547719, KJ547720, KJ547721, KJ547722, KJ547723, KJ547724, KJ547725, KJ547726, KJ594081, KJ594082, KJ594083. The integron sequences were also registered within the INTEGRALL database (http://integrall.bio.ua.pt/).24
The qacΔE1 gene was amplified using the primers 3′CSF2 TCAGGTCAAGTCTGCTT) and qacR (GGCACTGCATCCATGACG). The PCRs contained 1U Taq DNA polymerase (Invitrogen), 1× reaction buffer, 2.5mM MgCl2 (Invitrogen), 200μM dNTP (Biolabs) and 1μM of each primer in a total volume of 25μL. Amplification conditions were denaturing at 94°C for 5min, followed by 35 cycles of 94°C for 1min, 57°C for 1min, and 72°C for 2min, with a final extension step at 72°C for 10min. The PCR products were separated by gel electrophoresis on a 1% agarose gel in TAE buffer stained with ethidium bromide (0.5μg/mL) and visualized using UV transillumination. The strains of E. coli ATCC 25922 and P. aeruginosa blaVIM-1 were used as negative and positive controls, respectively.
Evaluation of minimal inhibitory concentration (MIC) for quaternary ammonium compounds (QAC)The MIC of QAC (10%, w/v, benzalkonium chloride, monoquaternary mixture of alkyldimethylbenzylammonium chloride) was determined by using the broth microdilution method25 with a concentration range of 14.75–118mg/L. The cultures at 105cfu/mL were added to each well of a microtiter plate containing 300μL Muller Hinton broth and different concentrations of QAC. Negative control used only Muller Hinton broth and positive control used broth with known bacterial inoculum. The assays were carried out in duplicate, for both samples and controls. The MIC was defined as the lowest concentration of the QAC at which complete inhibition of growth was observed at 24h and under optimal incubation conditions. The isolates were considered QAC resistant when the MIC was ≥59mg/L.25 To facilitate visualization of MIC, 20μL of a 1% solution of 2,3,5-triphenyl tetrazolium chloride (TTC) was added to each well of the microplate after the incubation period; E. coli ATCC 25922 was used as positive control.
ResultsIn a previous study, 518 colonies were isolated from m-Endo Les Agar and inoculated on Eosine Methylene Blue Agar resulting in 441 presumptive E. coli colonies.15 The intI1 gene was detected in 62 isolates and 43 that were resistant to two or more antimicrobials agents were selected for variable region of the class 1 integron analyses. ERIC-PCR was performed to assess the genetic diversity of the E. coli isolates that carried the class 1 integron and to exclude clones.
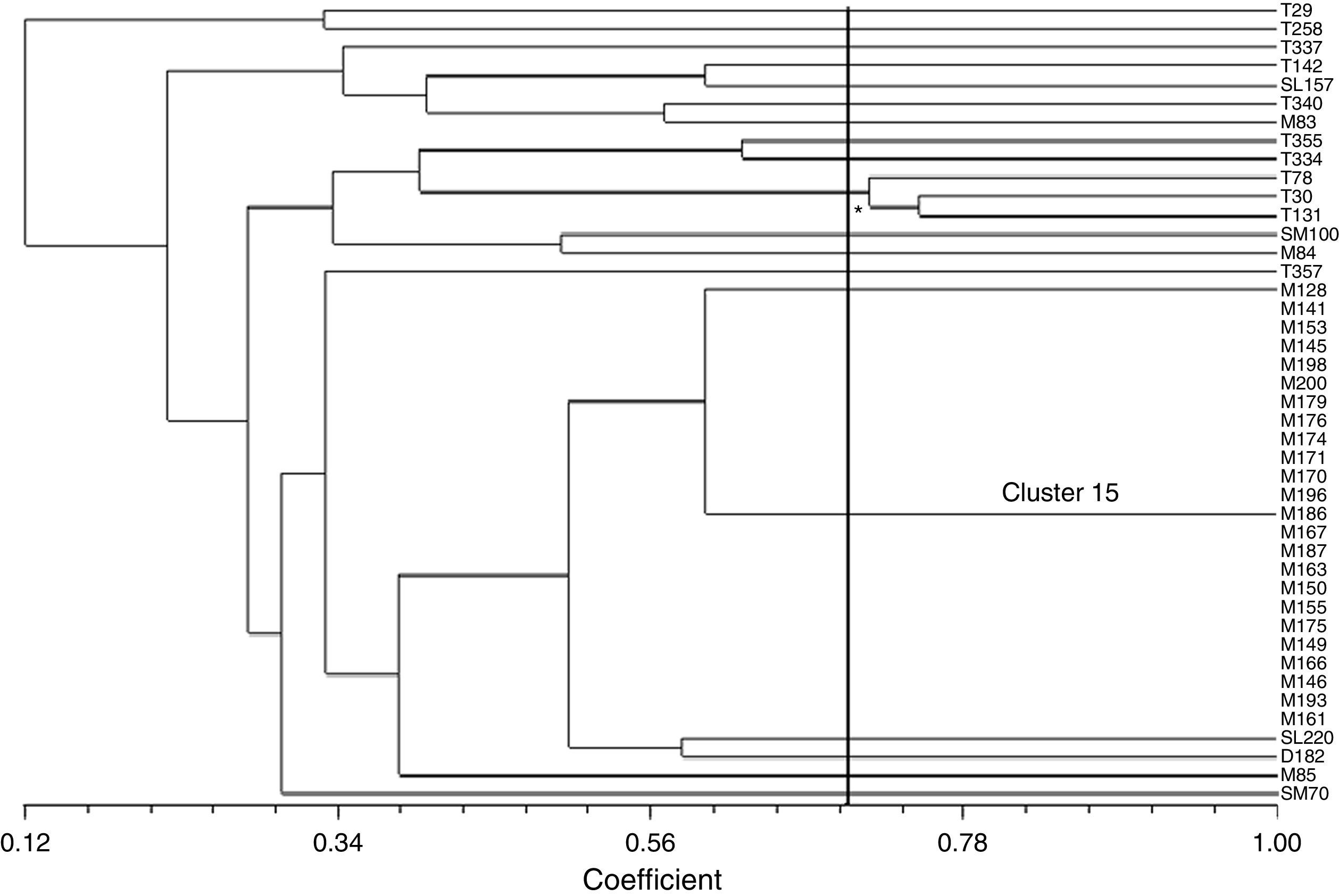
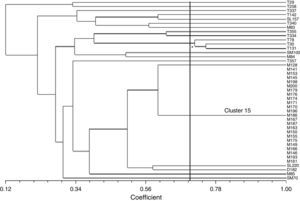
The isolates showed great genetic diversity and a dendrogram was constructed based on 43 isolates which generated 19 groups with a similarity of 70%. Most clusters were represented by a single isolate, except two clusters which comprised 3 (cluster 10) and 23 isolates (cluster 15). The largest cluster, cluster 15, comprising 23 isolates with 100% similarity and identical resistance pattern (AmSxTeChSmSu) were all from the same sampling site (Museum). As these isolates were considered clones, only 7 isolates were randomly chosen for analysis of the integron variable region. Cluster 10 was the second largest group and consisted of three isolates with 71% similarity; all from the same sampling site (Tapes). However, these isolates had a variable antimicrobial susceptibility pattern even though they shared resistance to tetracycline, spectinomycin, sulfonamide and trimethoprim–sulfamethoxazole, apart from resistance to other antimicrobials (ampicillin, norfloxacin, chloramphenicol and amikacin). These isolates also presented distinct integron gene cassettes (Fig. 2; Table 1). Contrarily, isolates M83 and M85, although from the same sampling site and with identical resistance pattern (AmSxTeChSmSu) and integron gene cassette, were genetically distinct with less than 50% similarity to cluster 15 (Fig. 2; Table 1).
Dendrogram for the ERIC-PCR pattern of E. coli isolates resistant to antibiotics with class 1 integron from superficial water calculated using the Jaccard coefficient and UPGMA clustering. Isolates T – city of Tapes; SL – city of São Lourenço do Sul; B, SM, M – estuary of Patos Lagoon. The axis indicates the similarity index and the vertical line indicates the 70% similarity cut-off. *Asterisk indicates cluster 10.
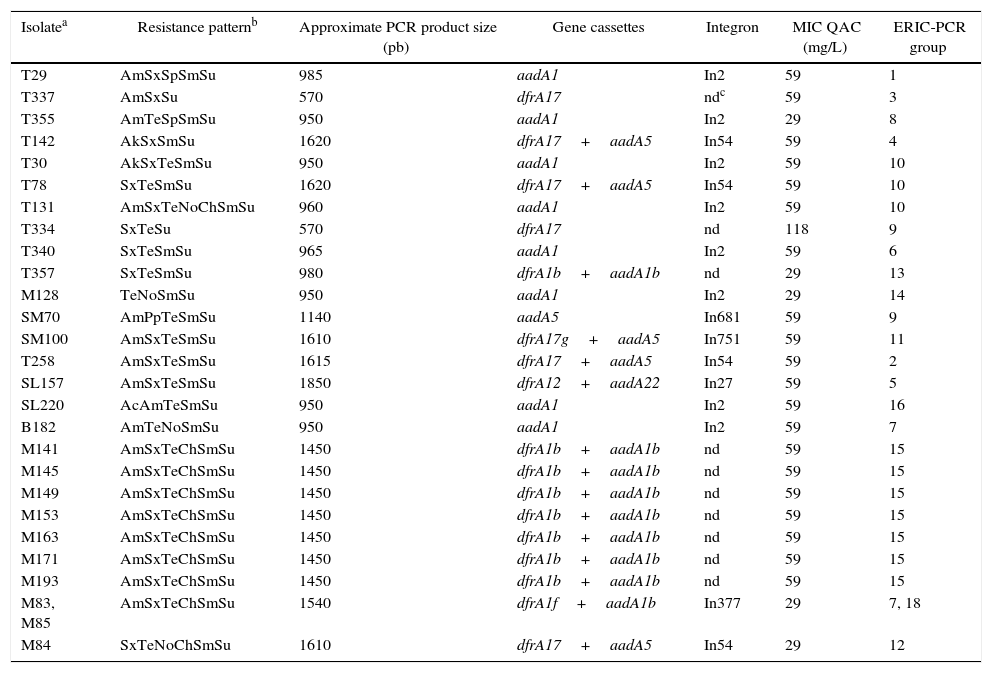
Resistance pattern, gene cassettes, integron, MIC quaternary ammonium and ERIC-PCR group in E. coli isolates with a variable region class 1 integron from the surface water of Patos Lagoon, Brazil.
| Isolatea | Resistance patternb | Approximate PCR product size (pb) | Gene cassettes | Integron | MIC QAC (mg/L) | ERIC-PCR group |
|---|---|---|---|---|---|---|
| T29 | AmSxSpSmSu | 985 | aadA1 | In2 | 59 | 1 |
| T337 | AmSxSu | 570 | dfrA17 | ndc | 59 | 3 |
| T355 | AmTeSpSmSu | 950 | aadA1 | In2 | 29 | 8 |
| T142 | AkSxSmSu | 1620 | dfrA17+aadA5 | In54 | 59 | 4 |
| T30 | AkSxTeSmSu | 950 | aadA1 | In2 | 59 | 10 |
| T78 | SxTeSmSu | 1620 | dfrA17+aadA5 | In54 | 59 | 10 |
| T131 | AmSxTeNoChSmSu | 960 | aadA1 | In2 | 59 | 10 |
| T334 | SxTeSu | 570 | dfrA17 | nd | 118 | 9 |
| T340 | SxTeSmSu | 965 | aadA1 | In2 | 59 | 6 |
| T357 | SxTeSmSu | 980 | dfrA1b+aadA1b | nd | 29 | 13 |
| M128 | TeNoSmSu | 950 | aadA1 | In2 | 29 | 14 |
| SM70 | AmPpTeSmSu | 1140 | aadA5 | In681 | 59 | 9 |
| SM100 | AmSxTeSmSu | 1610 | dfrA17g+aadA5 | In751 | 59 | 11 |
| T258 | AmSxTeSmSu | 1615 | dfrA17+aadA5 | In54 | 59 | 2 |
| SL157 | AmSxTeSmSu | 1850 | dfrA12+aadA22 | In27 | 59 | 5 |
| SL220 | AcAmTeSmSu | 950 | aadA1 | In2 | 59 | 16 |
| B182 | AmTeNoSmSu | 950 | aadA1 | In2 | 59 | 7 |
| M141 | AmSxTeChSmSu | 1450 | dfrA1b+aadA1b | nd | 59 | 15 |
| M145 | AmSxTeChSmSu | 1450 | dfrA1b+aadA1b | nd | 59 | 15 |
| M149 | AmSxTeChSmSu | 1450 | dfrA1b+aadA1b | nd | 59 | 15 |
| M153 | AmSxTeChSmSu | 1450 | dfrA1b+aadA1b | nd | 59 | 15 |
| M163 | AmSxTeChSmSu | 1450 | dfrA1b+aadA1b | nd | 59 | 15 |
| M171 | AmSxTeChSmSu | 1450 | dfrA1b+aadA1b | nd | 59 | 15 |
| M193 | AmSxTeChSmSu | 1450 | dfrA1b+aadA1b | nd | 59 | 15 |
| M83, M85 | AmSxTeChSmSu | 1540 | dfrA1f+aadA1b | In377 | 29 | 7, 18 |
| M84 | SxTeNoChSmSu | 1610 | dfrA17+aadA5 | In54 | 29 | 12 |
T, city of Tapes; SL, city of São Lourenço do Sul; B, Barra; SM, Saco da Mangueira; M, Museum located estuary of Patos Lagoon.
Among the 43 isolates of resistant E. coli studied here, the genetic content of the class 1 integron was analyzed in 27 isolates, of which 18 isolates were considered multiresistant. As shown in Table 1, the size of the variable region PCR product ranged between 0.570kb and 1.850kb and consisted of previously reported integron sequences such as In2, In54, In27, In377, In681 and In751. The isolate T357 and the seven isolates from cluster 15 did not display the complete sequence of the aadA1b gene cassette, and thus, these integrons could not be numbered, even if they were very likely to be identical to In377. A similar scenario occurred in isolates T337 and T334 which presented an incomplete sequence of the dfrA17 gene, and we hypothesize that this probably occurred due to an insufficient extension time (2min) used during PCR.
Gene cassettes encoding the dihydrofolate reductase genes (dfrA1, dfrA12 and dfrA17), which confer resistance to trimethoprim, and those encoding aminoglycoside adenyltransferase genes (aadA1, aadA5 and aadA22) responsible for resistance to streptomycin/spectinomycin, were also found among the isolates (Table 1).
The MICs for QACs ranged between 118mg/L and 29mg/L. While most isolates were resistant to QAC (MIC≥59mg/L), 36 isolates showed an MIC of 59mg/L, one isolate had an MIC of 118mg/L and 6 isolates had an MIC of 29mg/L; these 6 isolates were thus considered susceptible to QACs (Table 1). All the 43 isolates displayed the qacEΔ1 gene.
DiscussionERIC-PCR analysis revealed a high degree of diversity among the E. coli isolates, probably due to the different sources of contamination draining into the lagoon. There was low overall similarity (<40%) and clustering, indicating that the isolates from the Tapes sampling site were genetically distinct from those of the Museum sampling site. Nevertheless, the presence of isolates with 100% similarity, identical resistance patterns and integron gene cassette sequence indicate that either a clonal population is being released into the aquatic environment from a common source or that these strains might be persistent at the sampling sites. The latter scenario seems possible because it has been reported that some E. coli strains can persist in secondary habitats such as riverbank soils and temperate soils.26,27 Antimicrobial resistance attributable to the presence of integrons has been frequently reported in clinical E. coli isolates28 and in environmental isolates.12,29 Integrons promote the spread of antimicrobial resistance, not only within a species but also between species and genera.30 However, the prevalence of antimicrobial resistance in aquatic environments has received relatively little attention.
The occurrence of antimicrobial-resistant bacteria in aquatic environments in Brazil has considerably increased in recent years. A study on a hospital wastewater treatment plant in Rio de Janeiro, Brazil, detected ESBL (Extended Spectrum Beta Lactamases) and KPC (Klebsiella pneumoniae carbapenemase) producing Enterobacteriaceae in both untreated and treated sewage.31 Recently, Enterobacteriaceae and Aeromonas spp. KPC-producing were recovered from the sewage of a tertiary teaching hospital and also from a municipal wastewater treatment plant located in São Paulo, the largest city in Brazil.32 This report also showed that KPC-producing bacteria were present in the effluent of the wastewater treatment plant which was being discharged into an urban river.32 Additionally, KPC-producing K. pneumoniae has also been recovered from other urban rivers in São Paulo.33,34
Coutinho et al. (2014)35 analyzed the diversity of antibiotic-resistant bacteria occurring in aquatic environments subjected to distinct degrees of anthropogenic effects in the city of Rio de Janeiro and detected microbial communities capable of tolerating antibiotic concentrations up to 600 times higher than the clinical dose. Potentially pathogenic species were among the resistant organisms detected and some of them were multiresistant. Basso et al. (2014)36 report that polluted water from the Dilúvio stream (Porto Alegre, Southern Brazil) may act as a reservoir for the dissemination of antibiotic-resistance and enterotoxin producing Staphylococcus species into the community at large. Together, these studies demonstrate a high degree of antibiotic resistance among environmental bacteria in Brazil and provide evidence that pollution influences the observed diversity among drug-resistant bacteria in these aquatic habitats.35
In the environment, bacteria may exchange genes through horizontal gene transfer, which is one of the main mechanisms used to spread resistance traits. Aquatic habitats that receive wastewater discharge are hotspots for horizontal gene transfer; and in such sites, distinct bacterial communities from several sources come together, thus creating an ideal environment for gene exchange.35
In the present study, the variable region of the integrons showed little diversity and only genes encoding resistance to streptomycin, spectinomycin (aadA) and trimethoprim (dfr) were observed. Sequence analyses revealed the presence of six different integrons and In2 and In54 as most common. These two are also the more frequently found integrons among the E. coli strains in the INTEGRALL database, and are followed by In27, In377, and In751. Even though strains T258 and SM100 had an identical resistance pattern and showed integron PCR products that were very similar in size, sequence analysis revealed that they were very different integrons. The threshold used to distinguish between gene cassettes was a single amino-acid variation, thus two similar arrays of gene cassettes differing by only one amino acid would actually consist of different combinations of genes and are, therefore, assigned different In numbers. For example, while In54 designates reference dfrA17/reference aadA5, In751 indicates the dfrA17g allele/reference aadA5 (dfrA17g is reference dfrA17 with a Gly124Ala mutation).
Seven different gene cassettes were detected and eight strains showed a dfrA1b–aadA1b arrangement, while other eight displayed the aadA1 gene, and most of the isolates presented at least one kind of aadA cassette in the variable region (aadA1, aadA1b, aadA5 and aadA22), either alone or in combination with other cassettes. It is known that these two gene cassettes are commonly found in the integrons of E. coli isolated from various environments.3,37-39
The variable region of the class 1 integron was analyzed in 27 E. coli isolates and all the isolates were resistant to sulfonamide while 22 isolates were resistant to trimethoprim–sulfamethoxazole. Among these, 18 isolates had the dfrA12, dfrA17, dfrA17g, dfrA1b or dfrA1f gene in the variable region, corroborating previous observations that class 1 integrons are important for the spread of resistance to trimethoprim–sulfamethoxazole.40 On the other hand, four isolates with trimethoprim–sulfamethoxazole resistance did not display the dfrA genes in the variable region of the integron. A possible reason for this observation is that the strains probably carry larger integrons which were not amplified by the PCR conditions used in this study or that the sulI gene was present in the 3′conserved segment of the integron; the latter would be a key factor in determining resistance to trimethoprim–sulfamethoxazole and sulfonamide in these E. coli isolates.
We also observed that genes not associated with resistance to β-lactam antibiotics (ampicillin, piperacillin–tazobactam, amoxicillin–clavulanate), or other antibiotics (amikacin, tetracycline, chloramphenicol and norfloxacin) were present in the variable region of the class 1 integrons analyzed. This finding implies that resistance pattern need not always be linked only to integrons carrying antibiotic-resistance gene cassettes, and that other mobile genetic elements such as plasmids and transposons may also carry resistance genes.29,41,42 Plasmids carrying the ESBL genes are frequently conjugative and also carry determinants of resistance to non-beta-lactams such as tetracyclines, quinolones or aminoglycosides. Furthermore, we have previously observed that some of these isolates show a reduction in MIC for tetracycline and ampicillin in the presence of efflux pump inhibitors such as carbonyl-m-chlorofenilidrazone (unpublished data), reinforcing an idea that other mechanisms of antimicrobial resistance potentially collaborate to yield the phenotype of multidrug resistance.
Finally, a majority of isolates were resistant to QACs although they showed relatively low MIC, and all isolates displayed the qacEΔ1 gene. The qacEΔ1 gene is a typical component in the 3′-CS region of the class integron 1 and confers resistance to quaternary ammonium compounds; nevertheless class 1 integrons lacking this normal 3′-CS have also been reported.10,43 Quaternary ammonium compounds are commonly found in cationic biocide formulations; therefore, the use of various cationic biocides may also play an important but indirect role in the selection of these antibiotic-resistant bacteria.44 Contrarily, six isolates carrying this gene were susceptible to QAC; it has been reported that resistance to quaternary ammonium compounds is strongly affected by the conditions under which bacteria are exposed to QACs and that the mere presence of the qacEΔ1 gene does not necessarily imply resistance.45 For example, have demonstrated that the qacEΔ1 gene does not play an important role in resistance to benzalkonium chloride, a QAC, in P. aeruginosa.46 One explanation for this observation could lie in the fact that qac-mediated resistance targets a broad spectrum of distinct cationic compounds which in turn results in the exclusion of a high-level of specificity to certain QACs.47 Further, as mentioned above, it is also known that qac-mediated resistance could be affected by specific conditions under which the bacteria were exposed to the QACs.
In summary, this study characterized the variable region of the class 1 integron in antimicrobial-resistant E. coli strains isolated from an aquatic environment and compared their genetic profile to find antibiotic resistance patterns. It was found that the resistance pattern could not be entirely attributed to the majority of the genes present in the variable region. Nevertheless, integrons probably contribute to the spread of resistance and also possibly synergize with other mechanisms of resistance, thereby explaining the multiresistance patterns observed among the isolates. Environmental water habitats receive fecal contamination from different sources and are ideal environments for the dissemination of antimicrobial resistance among various species. The results of this study reflect the fact that the presence of the class 1 integron in fecal bacteria isolated from an aquatic environment can be considered a risk to public health as these bacteria are important reservoirs of antibiotic-resistance genes and can transfer these genetic determinants of resistance to other bacteria capable of infecting humans. The resistance to quaternary ammonium compounds is also of concern since these are often used in many biocidal formulations and may contribute to the selection of antibiotic-resistant bacteria in these environments.
Conflicts of interestThe authors declare no conflicts of interest.
This study was supported by CAPES (Brazilian Government Supporting Agency). The authors would like to thanks Dr. Thomas Jové (INTEGRALL curator) who kindly provided the information about the integrons.