The development of showerhead biofilms exposes the user to repeated contact with potentially pathogenic microbes, yet we know relatively little about the content of these aggregates. The aim of the present study was to examine the microbial content of tape-like films found protruding from a domestic showerhead. Culturing showed that the films were dominated by aerobic α- and β-proteobacteria. Three isolates made up almost the entire plate count. These were a Brevundimonas species, a metalophilic Cupriavidus species and a thermophile, Geobacillus species. Furthermore, it was shown that the Cupriavidus isolate alone had a high capacity for biofilm formation and thus might be the initiator of biofilm production. A clone library revealed the same general composition. However, half of the 70 clones analyzed could not be assigned to a particular bacterial phylum and of these 29 differed from one another by only 1–2 base pairs, indicating a single species. Thus both the culture dependent and culture independent characterizations suggest a simple yet novel composition. The work is important as the biofilm is fundamentally different in form (tape-like) and content from that of all previously reported ones, where variously Mycobacterium, Methylobacterium and Xanthomonas species have dominated, and extremophiles were not reported.
Biofilms are microbial communities that grow attached to surfaces and/or interfaces; they are embedded in a frequently self-produced matrix of extracellular polymeric substances.1 The development of biofilms in bathroom environments brings the microbes present into close contact with humans. Biofilms have been reported in washstands,2 on shower curtains3 and in showerheads.4,5 They can constitute potential reservoirs for pathogens, particularly for immune-compromised individuals. Showerhead biofilms have, for example, been shown to include and enrich pathogenic (e.g., Legionella pneumophilia, Mycobacterium avium) and opportunistic pathogens such as non-tuberculous mycobacteria.4 Microbes which dislodge from shower biofilms come into contact with the skin and also the pulmonary system by way of aerosolization. It has been hypothesized that the recent rise in pulmonary infection with non-tuberculous mycobacteria is linked to the increased use of showers rather than baths.6 Showerheads can be considered as extreme environments. For example, microbes present in mature biofilms must survive at least short periods of high temperatures, and bathroom biofilms might then enrich for thermophilic species. Most studies have focused on the specific presence of Legionella pneumophilia and Mycobacterium avium, and little is known of the prevalence and identities of other microorganisms that may be delivered during shower usage. In fact, to the best of our knowledge there exist only two comprehensive investigations of showerhead biofilms.4,5 A related study of shower curtain biofilms has also been published.3 In one study,4 it was reported that the showerhead environment strongly enriched for microbes able to form biofilms in water systems, including Methylobacterium spp. and Sphingomonas spp. and, most strikingly, non-tuberculous mycobacteria which accounted for more than about one third of the clone library. As the study was non-culture based and did not include metabolic stains, it did not provide information on the metabolically active and culturable fraction of the analyzed biofilms. Strikingly different were the results of pyrosequencing of 16S rRNA genes amplified from biofilms on the outer surface (spray plate) of three showerheads.5 A preponderance of γ-proteobacteria was demonstrated, with the genus Xanthomonas accounting for more than one third of the total sequence information. The corresponding culture-based analysis showed a much more even distribution of a wide-range of gram-negative and gram-positive genera. Given the small number of investigations, the detection of potentially pathogenic bacteria and the wide variation in the results found, there is need for more studies to improve the field. There is great variation in the origins (e.g., surface and groundwaters), temperature and quality of domestic water supplies, as well as in water disinfection practices. We thus hypothesize that as more studies of showerhead biofilms become available, new and strikingly different contents of potential health significance might be revealed. Previous studies have collected biofilms by either swabbing or scraping, and no mention is made of observed variations, if any, in the appearance of biofilms collected. The biofilm analyzed in the present work was unusual in that it presented as free-hanging, tape-like strands which must at least in part be attributable to the properties of the species present in it. The present study uses both culture and non-culture based techniques, to identify the composition of the biofilm. Unlike previous work it also looks at the content of fungi and protozoa and investigates the potential for biofilm formation among individual bacterial isolates. The latter may help to identify the main progenitors of biofilm formation and aid in the development of strategies to control biofilm development.
Materials and methodsBiofilm origin, appearance and collectionThe biofilm consisted of strips (>1cm) of slightly opaque, elastic, tape-like material which had emerged and hung from the vents in a domestic, plastic showerhead. The showerhead was delivered to a local Norwegian water works by a concerned resident who found the material unsightly. The home received UV and hypochlorite treated surface water from a municipal water works. Small pieces of the material were aseptically sectioned with a scalpel and washed carefully with 5 serial changes (10ml aliquots) of pharmaceutical grade, sterile distilled water prior to analysis.
Live/dead cell staining and epifluorescence microscopyTo determine its general structure, content and metabolic state, washed biofilm sections were stained using the FilmTracer™ LIVE/DEAD® Biofilm viability kit (Invitrogen, CA, USA). In brief, 3μl SYTO® 9 green fluorescent stain and the same volume of propidium iodide (red fluorescence) was added to 1ml filter sterilized water (the stain). The stain was pipetted onto biofilm sections which were kept in the dark. After 30min. the residual stain was removed by two washes in pharmaceutical grade water. The biofilm sections were examined by fluorescence microscopy (Olympus BX40, GmbH) using the antifadent containing immersion oil Citifluor AF87 (Citifluor Ltd, London, UK) and emission filter sets for the green (WIBA-cube, Olympus) and red (WG-cube, Olympus) dyes.
Plate count investigation of the biofilm content (bacteria, fungi, protozoa), and cellular morphologiesWashed biofilm (10mg) was added to 1ml of pharmaceutical grade distilled water in an eppendorf tube. Thereafter, 1g of acid washed, sterile 2mm glass beads were added to the tube which was then vortexed at maximum speed for 2×5min with intermittent cooling (2min) on ice. This approach seemed to completely disrupt the film and free bacteria were visible in the microscope. Aliquots (0.1ml) were spread on R2A-agar (Oxoid, Thermo Scientific, UK) to obtain the total mesophilic heterotrophic bacterial plate count and Rose Bengal Chloramphenicol agar (Oxoid) to obtain the total mesophilic heterotrophic fungal plate count. Plates of each agar type were incubated at 22±2°C (7 days). An additional R2A plate was incubated at 55±1°C for 48h to detect thermophiles. Samples were also spread on sheep blood agar plates (Oxoid) for the detection of rapidly growing strains of potential clinical interest. Plates were examined for colony counts and types after aerobic and anaerobic incubation at 37±1°C (48h).
Free-living protozoa were detected as previously described.7 In brief, washed biofilm pieces (10mg) were added to non-nutrient agar seeded with heat-killed Escherichia coli. Plates were incubated at two different temperatures, 22±2°C and 37±1°C, and examined for the presence of protozoa and protozoal cysts over a period of 7 days.
DNA-isolation from the biofilmExtraction of total community DNA from biofilm pieces was performed using the alternative protocol in the UltraCleanTM Soil DNA Isolation kit (MoBIO Laboratories, CA, USA). In brief, 10mg washed biofilm was first transferred into the supplied DNA extraction tubes. As an aid to homogenization of the material, the tube contents were mixed for 10min at 250rpm prior to the DNA isolation steps. The concentration of the cleaned DNA was measured with a nanodrop (Saveen Werner, Sweden) and the DNA was stored at −80°C until use.
Colony-polymerase chain reaction (colony-PCR) of the 16S and 18S rRNA-genes and sequencing studiesThe genetic identity of bacterial biofilm isolates was investigated by ribosomal DNA typing of part of the 16S rRNA-gene using the primer pair 341F (5′-CCTACGGGAGGCAGCAG-3′) and 907R 5′-CCGTCAATTCMTTTGAGTTT-3′ and a pinhead of colony material as the source of DNA exactly as previously described.8 The primers bind to highly conserved regions of the bacterial 16S rRNA gene to provide PCR products of approximately 590bp. In some instances, where good identifications were not obtained, the primer pair 27F/1492R was also used to provide a longer amplicon for sequencing.9
Fungal isolates were identified as previously described7 by sequencing the product obtained using the eukaryotic 18S ribosomal DNA primer set Euk1209f and Uni1392r which give a PCR product of about 200bp.
Sequencing reactions were performed by a commercial laboratory (ABI-lab, University of Oslo, Norway) using the forward and reverse primers used in production of the PCR products. Poor quality data and primer sequences were removed from the sequence terminals prior to similarity searches. Sequences were analyzed and assigned to taxa using the ‘Classifier’10 function with an 80% confidence threshold cutoff. The ‘16S rRNA training set’ for bacteria and the ‘UNITE Fungal ITS trainset 07/04/2014’ for fungi were referenced. Both sets are available at the ribosomal database (RDP) site.11 For the production of pie charts, phylum- and class-level (for the Proteobacteria) designations were employed.
Cupriavidus species-specific PCR and metal ion susceptibilityA species-specific PCR using primers designed to target the signal transduction histidine kinase gene of C. metallidurans was performed exactly as previously described.12 For the determination of metal ion susceptibility, the Cupriavidus sp. isolated from the biofilm, and the control strains Pseudomonas aeruginosa (DSM 1128) and Serratia marcescens (ATCC 14756), were inoculated onto R2A agar (pH=7.2) amended with either Co2+, Cu2+ or Zn2+ ions in doubling dilutions in the concentration range 0.63–10mM. Plates were examined for growth (presence/absence) over a period of 14 days incubation at 28±1°C.
Cloning and sequencing of biofilm community DNACloning and transformation was performed using a TOPO TA Cloning Kit (Invitrogen). In brief, 0.5–1.5μl of PCR product per μl vector were ligated as specified in the kit's complete protocol. PCR was performed exactly as described for colonies (primers 341F/907R), except that biofilm DNA (0.5–3μl; 30ng/μl) was used as template. Isolated plasmids were checked for the presence of an insert of correct size prior to sequencing, by restriction analysis using EcoR1 which cuts on both sides of the linker region. Clones (70 in total) were randomly selected for sequencing with the vector-specific primers M13F and M13R supplied with the kit. Primer sequences were removed before similarity searches (see above) were performed.
Biofilm formation potentialBiofilm formation assays were performed following a previously described method13 with a few modifications. In brief, a small amount of a single colony on tryptone–soya agar (Oxoid) was used to inoculate 5ml tryptone–soya broth (Oxoid). Tubes were shaken at 100rpm at 22±2°C or 55±1°C (for thermophiles) for 48h. The growth was collected by centrifugation (5000rpm) and the supernatant was discarded. The pellet was homogenized in fresh TSB such that OD595=0.15. Thereafter, 200μl suspension was added in triplicate to 96-well flat bottom polystyrene tissue-culture treated plates (Costar® cat. nr. 3628; Corning Life Sciences, MA, USA). Some wells contained only TSB as controls. The plates were lidded and sealed with parafilm before being set to shaking at 100rpm at the above mentioned temperatures. After 30h, the culture OD595 was read directly in the wells using a Victor multilabel counter (Perkin Elmer, Boston, MA). Using a multichannel pipette the liquid was carefully removed, taking care not to touch the biofilm and avoiding bubble formation. Biofilms were washed, stained and solubilized exactly as described in the referenced study. Based on the OD595 measurements, ‘biofilm formation’ (BF) and ‘specific biofilm formation’ (SBF, which normalizes BF against the amount of planktonic growth) were measured and categorized according to several different formulae given in the literature. Three previously published, widely-used formulae designed for this purpose were used. Two of these are described with original references in the paper by Naves and others.14 In brief:
where, AB=OD of stained attached bacteria; CW=OD of stained control wells; G=OD of cells grown in suspended culture.Based on testing of a large number of strains, Naves and others proposed the following scoring system (see Table 1) expressing the tendency to forms biofilms.14 This system is also adopted in the present study.
Semiquantitative classification of biofilm production using three different formulae (above) and category cut-off values.
| Formula | Strong (S) | Moderate (M) | Weak (W) | Negative (N) |
|---|---|---|---|---|
| BF=AB/CW | ≥6.00 | 4.00–5.99 | 2.00–3.99 | <0.200 |
| SBF=(AB−CW)/G | ≥1.10 | 0.70–1.09 | 0.35–0.69 | <0.35 |
| BF relative to OD.c | ODbf>4OD.c | 2OD.c≤ODbf≤4OD.c | OD.c<ODbf≤2OD.c | ODbf≤OD.c |
A third and fundamentally different approach to express BF is that described by Stepanovic and others.15 Here 4 classes are defined relative to the negative control using a defined cut-off optical density (OD) value=(OD.c). OD.c is defined as being three standard deviations above the mean optical density of the negative control. The mean OD value of stained control wells (TSB, no cells) is subtracted from the mean OD of test wells giving the biofilm (bf) OD, ODbf. Using this approach the propensity to form biofilms is categorized as given in Table 1. This system was also applied to the biofilm isolates obtained in the present study. All tests were performed at least twice.
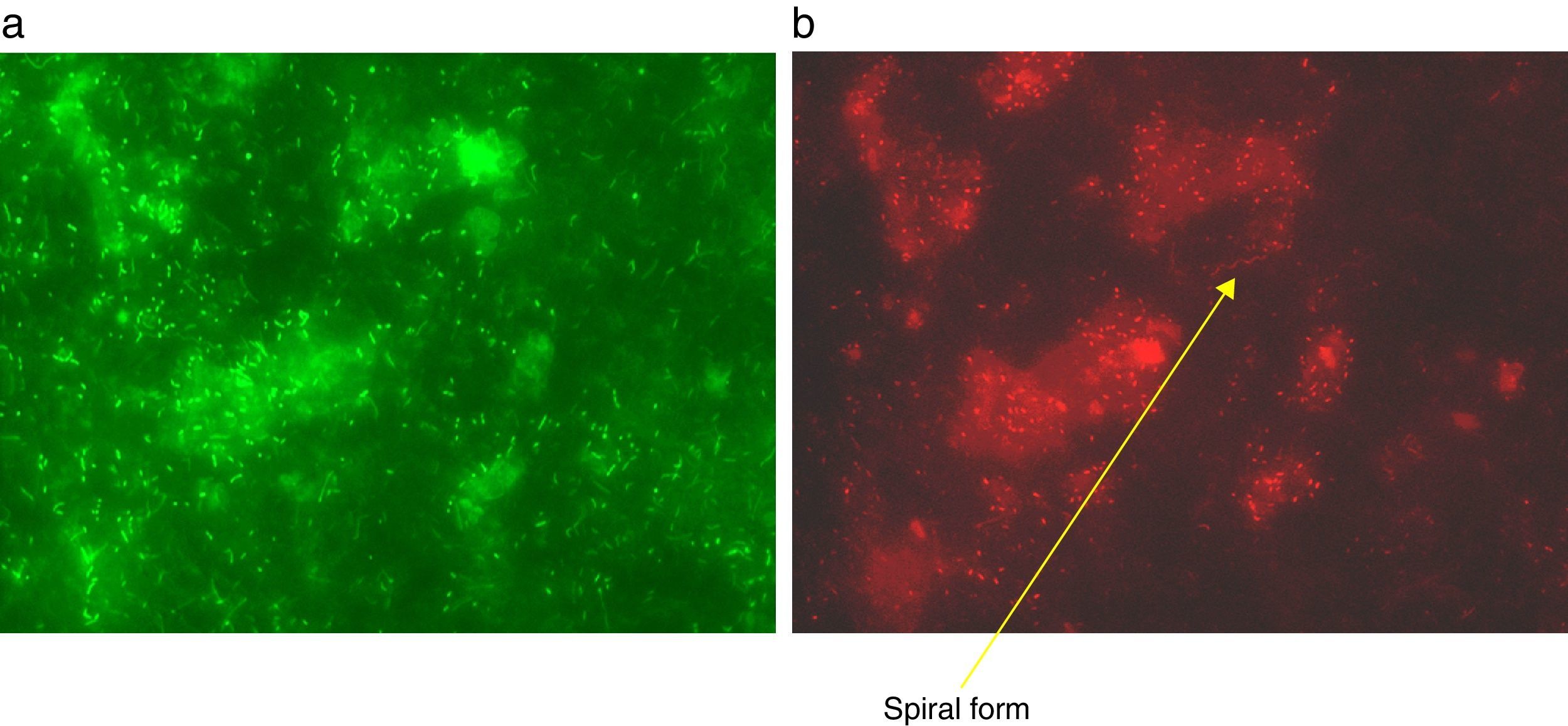
ResultsLive/dead cell staining and epifluorescence microscopyIn order to investigate if the harvested material was a true biofilm and to gain information on its general content and metabolic state, vital staining was employed. Examination of film sections by epifluorescence microscopy showed that the material was a bed of microbes spread in the film matrix. A variety of cellular morphologies with a clear predominance of rods and some spiral forms were visible (Fig. 1A, B). The large populations of both dead (red; membrane-damaged) and live (green; membrane intact) cells showed that the biofilm is at least in part a dynamic structure.
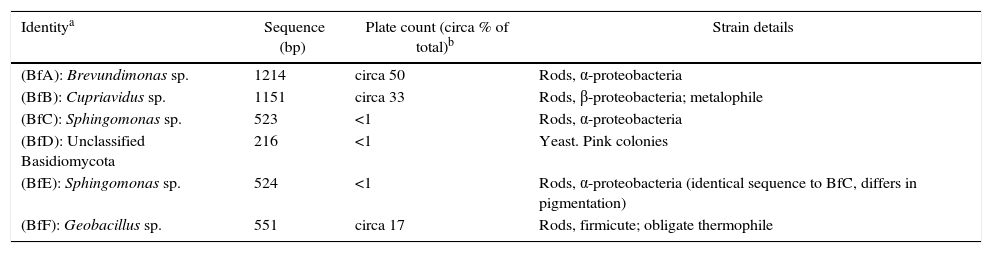
Plate count investigation of biofilm content (bacteria, fungi, protozoa) and cell structureIncubation on R2A for 7 days at 22±2°C produced a colony count of circa 104colony forming units (CFU)/mg homogenized biofilm. Based on colony morphologies, two strains accounted for circa 60% (hence designated Biofilm A, Bf A) and circa 40% (Bf B) respectively of the total mesophilic viable count. Smaller numbers of Bf C, D (a single colony, and the only fungal isolate) and E were detected. Plating of samples aerobically on blood agar (37±1°C; 48h) gave only a single, non-hemolytic colony type, which sequencing studies later showed to be identical with BfB. No colonies grew on blood agar incubated anaerobically. An additional R2A plate incubated at 55±2°C produced a population of circa 3×103CFU/mg biofilm of a single, thermophilic, strain, BfF. No fungal plate count was obtained on Rose Bengal Agar and no protozoa grew from the biofilm sections. Standard tests showed that the bacterial biofilm isolates were gram-negative rods, except for BfF which was a sporulating gram-positive rod. Thus, under the conditions tested, the culturable fraction of the biofilm was almost entirely composed of three species of aerobic rods (Table 2). The results are in accordance with the large number of rods seen in the vital-stained biofilm sections (Fig. 1A and B).
Identity and properties of the cultured fraction.
| Identitya | Sequence (bp) | Plate count (circa % of total)b | Strain details |
|---|---|---|---|
| (BfA): Brevundimonas sp. | 1214 | circa 50 | Rods, α-proteobacteria |
| (BfB): Cupriavidus sp. | 1151 | circa 33 | Rods, β-proteobacteria; metalophile |
| (BfC): Sphingomonas sp. | 523 | <1 | Rods, α-proteobacteria |
| (BfD): Unclassified Basidiomycota | 216 | <1 | Yeast. Pink colonies |
| (BfE): Sphingomonas sp. | 524 | <1 | Rods, α-proteobacteria (identical sequence to BfC, differs in pigmentation) |
| (BfF): Geobacillus sp. | 551 | circa 17 | Rods, firmicute; obligate thermophile |
The identities of the biofilm isolates based on sequencing studies and using the RDP classifier with the default ≥80% confidence threshold cutoff are given in Table 2. The cultured fraction (BfA–F) of the biofilm is heavily dominated by aerobic Proteobacteria. With the exception of BfB (β-proteobacteria) all proteobacteria were members of the α-class. Approximately half of the plate count was a Brevundimonas sp., whereas the other half was made up of two extremophiles: a Cupriavidus sp. (shown to be a metalophile, see below) and a Geobacillus sp. (a thermophile).
Cupriavidus sp.: species-specific PCR and metal ion susceptibilityThe metalophile Cupriavidus metallidurans is the best characterized member of the genus. C. metallidurans-specific-PCR produced no product, suggesting BfB is not this species. However, BfB possessed a metalophilic character and was much less susceptible to metal ions than the control strains Pseudomonas aeruginosa (DSM 1128) and Serratia marsecens (ATCC 14756). None of the strains tested were greatly restricted by Zn2+ in the range (0.63–10mM) investigated. No growth of the control strains was obtained above 0.63mM Cu2+ or Co2+, whereas BfB grew weakly at 2.5mM Cu2+. Strikingly, after adaptation to 0.63mM Co2+ (initial weak growth), the BfB grew at the highest Co2+ concentration (10mM) tested.
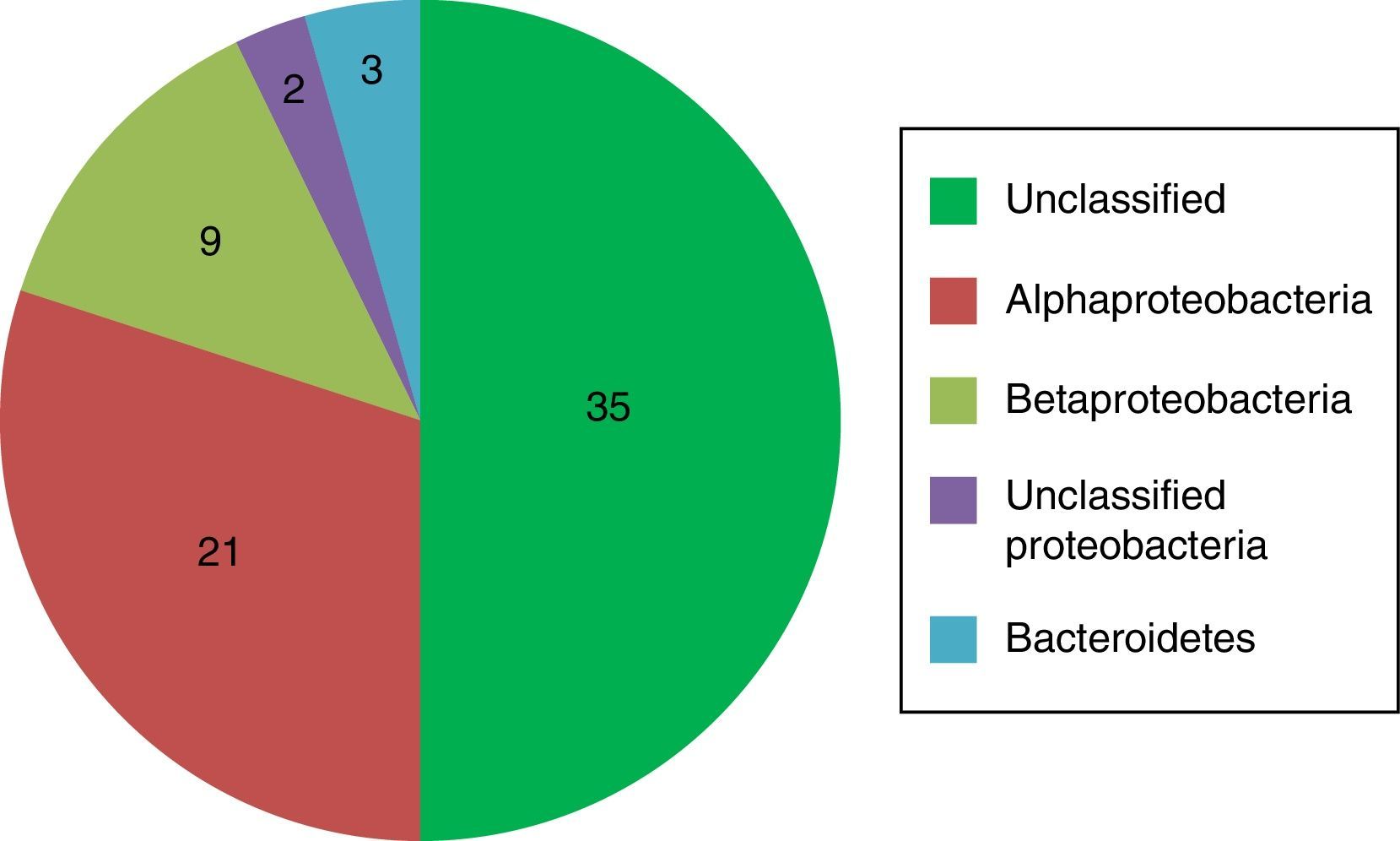
DNA-isolation and cloning and sequencing of biofilm community DNAA yield of about 30ng/μl of high quality community DNA was obtained from 10mg biofilm sections. After cloning and transformation of PCR-products, 70 clones were randomly selected for sequencing. Of the 70 clones chosen, 27 were unique DNA sequences. The library was dominated by 29 clones which differed by only 1–2bp across the entire sequenced length of 540bp (>99% similarity) and of which 20 were identical. This group was not assigned by the RDP classifier to any taxon at the phylum level, and thus a major portion of the biofilm cannot at present be identified based on the data generated by the study. Most of the other clones in the library represented α- and β-proteobacteria, and included both Brevundimonas, Cupriavidus and Sphingomonas DNA (also found in the cultured fraction), as well as the genera Caulobacter, Sphingobium, Phenylobacterium, Afipia, Bradhyrhizobium and Aquabacterium. The pie chart in Fig. 2 illustrates the division of taxa comprising the biofilm generated from total biofilm DNA. The numbers of clones of each taxon are indicated in the figure.
Biofilm formation potentialBrevundimonas sp., Cupriavidus sp. and Geobacillus sp. which made up about 99% of the readily culturable fraction of the biofilm, were tested for their ability to form biofilms. Three formulae (BF, SBF and BF as multiples of OD.c) which assign the propensity to form biofilms to 4 categories (Table 1) were applied. Using these guidelines BfA (BF=1.70±0.21; SBF=0.23±0.07; ODbf(0.11)≤OD.c(0.18)) and BfF (BF=1.12±0.006; SBF=0.006±0.001; ODbf(0.004)≤OD.c(0.038)) always scored as negative/non-producers of biofilm. However, BfB always scored as a ‘strong producer’ (BF=20.5±0.24; SBF=5.92±0.07; ODbf (3.17)>4OD.c (0.72)). Furthermore it was noticed that this strain formed a thick pellicle on TSB within 48h growth.
DiscussionThe present study uses both culture dependent and culture independent methods to determine the structure of an unusual, tape-like showerhead biofilm recovered from a Norwegian home. Using a similar approach to other groups,3,4 epifluorescence microscopy and vital staining showed that the isolated material was a true biofilm with a metabolically active fraction (Fig. 1A and B). This result was supported by the production of plate counts from homogenized biofilm sections. Although, as discussed below, no primary pathogenic species were grown from the biofilm, the plate-count revealed genera such as Sphingomonas and Brevundimonas which are known to include opportunistic pathogens of immune-compromised hosts.3,16 Thus, the biofilm might represent a health issue, particularly with respect to shower users with poor immune status.
The present study is different from previous work on showerhead and shower curtain biofilms,3–5 in that it looks not only at the bacterial content, but also at the fungal and protozoal fractions, as well as the content of thermophiles. Testing for thermophiles is logical as microbes present in showerhead biofilms must at least tolerate intermittent periods of high temperature. Furthermore, the study is novel in that it provides information on the tendency of the cultured strains isolated from the showerhead to form biofilms, which is an aid in understanding biofilm formation and control. However, the main importance of the present work is that it describes a showerhead biofilm with a radically different structure and bacterial composition than those revealed in previous studies. For example, swabs taken from the inner surface of 52 showerheads showed that 49.8% of the generated clones represented non-tuberculous mycobacteria or Methylobacterium spp.4 Neither of these were found in the present study. Similarly, pyrosequencing based on swabbing of the outer surface of 3 showerheads, showed that 50.3% of the sequences were derived from γ-proteobactera.5 This class was not found in the present work. The identified proteobacterial fraction belonged to the α- and β-proteobacteria classes (Table 2; Fig. 2). Both previous showerhead studies used the basic local alignment search tool, BLAST,17 to identify sequences to at least the level of closest well-characterized genus. Using this approach, neither study seems to report any sequences as ‘unclassified’. The more stringent approach to sequence identification adopted in the present study (i.e., use of the RDP classifier with a ≥ 80% confidence threshold cutoff) resulted in non-assignment of about one half of the clone sequences at the phylum-level (Fig. 2). Of the non-assigned sequences 41% showed >99% similarity across the entire sequenced length. Thus, a major portion of the biofilm may consist of a single unassigned species. The relatively large proportion of Proteobacteria is the major similarity between the present and previous work on showerhead and shower curtain biofilms.3–5 The finding of many Proteobacteria is perhaps not surprising, as the phylum includes many genera which are diverse in physiology and ubiquitous to water-environments.3
There were some similarities (e.g., the presence of α- and β-proteobacteria), and some notable differences between the biofilm compositions revealed by culturing and cloning/sequencing. This is generally speaking the expected result, as only a small proportion of the species present in complex environmental samples grow under standard conditions, - an observation which has been referred to as the ‘great plate count anomaly’.18 The major difference was the high proportion of unassigned sequences in the clone library which originate from a species not found in the cultured fraction. Explanations for this could be that the species in question did not grow under the culture conditions or it made up part of the dead cell fraction of the biofilm (Fig. 1b). About 17% of the total plate count (Table 2) was made up of the thermophilic Geobacillus sp., which was not found in the clone library. This could be explained by the presence of this species as endospores from which DNA is not readily extractable. Alternatively Geobacillus DNA may not have been detected due to low coverage of the clone library. The presence of thermophiles might be a pertinent example of the shaping of biofilms by abiotic factors, in this case high water temperatures. A survey of the literature suggests Geobacillus spp. are non-pathogenic.
Previous studies on showerhead biofilms made no attempt to study the native content of fungi and protozoa, and by focusing on bacterial identification only, may have overlooked small eukaryotes of health or ecological significance. Notwithstanding, with the exception of a single fungal colony which grew on the R2A plates, fungi and protozoa (including cysts) were not found. Using the present approach, the fungus could only be identified to the phylum level as an ‘unclassified Basidiomycota’ (Table 2). Although some members of this phylum are considered opportunistic pathogens (e.g., Cryptococcus neoformans), the presence of one or a few cells in the biofilm is unlikely to be a health-issue. However, we suggest that subsequent work on showerhead biofilms should include analyses for fungal content as a number of fungal genera found in domestic water are considered opportunistic pathogens.19 The rationale for screening for protozoa comes from the concern that free-living amoebae in water distribution systems can be Trojan horses for intracellular pathogens such as Legionella pneumophilia. In addition, some amoebae such as Acanthamoebae are important eye pathogens. Although protozoa were not detected, we have previously7 found using the described methodology, Acanthamoeba castellani and other amoebae in Norwegian drinking waters and, more relevantly, also in Norwegian domestic tap biofilms.20
Partial sequencing of the 16S rRNA gene strongly suggested that BfB is a Cupriavidus sp. Although the demonstrated metalophilic character of the isolate is typical for C. metallidurans, a species-specific PCR gave no product. BfB gave a MICCo2+>10 mM (highest concentration tested); whereas the MICCu2+ was 5.0mM. These values are similar to those reported for C. metallidurans.21,22 However, in an amended taxonomy of the genus, it is reported that resistance to various metals is widespread among Cupriavidus spp.23 For example, in addition to C. metallidurans, C. necatar is also known for its resistance to copper.24 However, a search of the available literature suggests that no serious health issues seem to be associated with any of the Cupriavidus species, so the main interest of the isolate remains its presence in and possible contribution to the formation of biofilms (see section on biofilm formation below). It is not known if a metalophilic character has significance for survival in the showerhead. Given the high water temperatures, leaching of metal ions from pipes might make the trait advantageous. The detection of Cupriavidus in a showerhead biofilm seems to be a completely novel finding.
To address the ability of the isolates to form biofilms, a semi-quantitative static model which has been demonstrated to be a reliable and reproducible method for assessing biofilm formation in vitro was used.13,14,25 Following the advice and approach of others,25 a number of formulae (3 in total, see materials and methods and Table 1) were used on the raw data to offset variations in culture conditions, methodologies and environmental factors. Of the plate count isolates, only the Cupriavidus sp. (strain BfB) showed a propensity to form biofilms and it was classified regardless of the formula used as a ‘strong’ producer. C. metallidurans has previously been shown to grow efficiently in biofilms, and it has been reported that even acid treatment failed to remove it from sand particles.21 Furthermore, the complete genome of the C. metallidurans type strain CH34 has been shown to contain many genes required for the biogenesis of fimbriae important for cell adhesion, colonization, and biofilm formation.26 No information on biofilm formation by other members of the genus seems to have been reported, so it is not known if a strong tendency to form biofilms is typical for Cupriavidus spp. It was also observed that strain BfB produced a thick pellicle on TSB broth. Pellicles can be considered air-liquid biofilms, whose formation consists of attachment to the culture vessel, production of a mono-layer and subsequent thickening to a three dimensional structure involving extracellular material.27 It is possible that the tape-like biofilm here described originated as a special case of pellicle formation by strain BfB and that this pellicle subsequently became colonized by the other biofilm members producing the mature biofilm. It will be interesting to see if future work shows C. metallidurans or other Cupriavidus spp. to be important contributors to the production of domestic biofilms, and if a metalophilic character has relevance in this respect.
Conclusions and suggestions for further workDespite its implication as a potential source of disease, the microbial composition of the showerhead environment is poorly known.4 The present study confirms that there can great variation in showerhead biofilm structure and content. Indeed amongst the published studies, variation rather than accord seems to be the rule. This indicates the need for more studies. The tape-like structure and content of extremophiles are novel with regards to showerhead biofilms. The study is also unique in its categorization of biofilm formation capacity among the cultured strains. This is seen as a logical first step for investigating how biofilms might arise. Recently it has been demonstrated that bacteria grown from biofilms on the outer surface of showerheads can coaggregate and autoaggregate.5 Such aggregates can facilitate biofilm formation, and similar tests could in future work be combined with measures of biofilm forming capacity, in a multi-faceted approach to the study of biofilm formation in the domestic environment.
Accession numbersAll DNA sequences have been assigned GenBank accession numbers: KJ540983–KJ541025.
Conflicts of interestThe authors declare no conflicts of interest.