DNA genotyping of Mycobacterium tuberculosis has been widely applied in the understanding of disease transmission in many countries. The purpose of this study was to genotype the strains of M. tuberculosis isolated in patients with new tuberculosis (TB) cases in Minas Gerais, as well as to compare the similarity, discriminatory power, and agreement of the clusters between the IS6110 Restriction Fragment Length Polymorfism (RFLP) and 12 loci Variable Number Tandem Repeat – Mycobacterial Interspersed Repetitive Units (MIRU-VNTR) techniques. It was observed that 32% (66/204) of the isolated strains in the RFLP-IS6110 and 50.9% (104/204) of the isolated strains in the MIRU-VNTR presented a similarity of equal to or above 85%. The RFLP-IS6110 and MIRU-VNTR proved to contain a high discriminatory power. The similarity index resulting from the RFLP showed no recent transmission. Good agreement was observed between the techniques when clusters were detected; however, the best epidemiological relationship was found when using the RFLP-IS6110.
Regarding countries in the world with tuberculosis (TB), Brazil is still ranked 18th in total of 22.1 In Minas Gerais (MG), in 2014, 3567 new TB cases were identified, reaching an incidence coefficient of 17.2 cases/100,000 inhabitants and 289 deaths. In 2007, the year in which the II National Survey of Drug Resistance was released, some 4810 new TB cases were identified, reaching an incidence coefficient of 25/100,000 inhabitants and 276 deaths. The cure percentage found within the new TB cases reached 56%, the patient's abandonment of treatment reached 7.2%, and the proportion of primary Multidrug-Resistant Tuberculosis (MDR-TB) was 1.8%.2 In this scenario, the treatment and discovery of new TB cases are as important as the investigation concerning the genetic diversity of the clinical isolates of Mycobacterium tuberculosis, which can be useful in gathering information on the origin and transmission of circulating strains.
DNA genotyping of M. tuberculosis, aimed at investigating the epidemiological relationships among TB cases, has been widely applied in many countries since the DNA typing technique was developed in the 1990s.3,4 The Restriction Fragment Length Polymorfism (RFLP-IS6110) revolutionized studies regarding the transmission of M. tuberculosis. The analysis of the band patterns of RFLP-IS6110 requires the use of expensive software and specialized professionals, and are quite difficult to use.5
Its advantage, however, is its genetic stability.6 The variable number of repetitions in sequence, the Variable Number Tandem Repeat – Mycobaterial Interspersed Repetitive Units (MIRU-VNTR) 12 and 24 loci was favorably evaluated and proposed as the new standard method for the analysis of M. tuberculosis.7 Its advantages are related to the need for a small quantity of DNA, given that this technique is based on the Polymerase Chain Reaction (PCR), with greater practicality and simplicity. Its disadvantage is related to genetic instability due to the silent mutations that can quickly change the genetic profile of the bacteria.8,9
The state of MG has the second largest population in Brazil, with regions in which the incidence of TB can reach 115/100,000 inhabitants. Added to this variation in the disease incidence is the size of this territory, the large number of cities, and the variations in socioeconomic conditions, which hinder the control of the disease.
The application of methods of molecular typing has advanced significantly in the understanding of the transmission and/or pathogenic properties of the mycobacteria, and provide new and powerful tools to combat and protect against the disease.10 Therefore, in MG, the study of molecular methods is still relatively unexplored, and their uses have become necessary to gain a better understanding of the genetic profile, transmission route, and dissemination of TB in this region. This study aims to genotype the strains of M. tuberculosis isolated in patients with new TB cases in MG and compare their similarities, discriminatory power, and agreement of the clusters between the RFLP-IS6110 and MIRU-VNTR 12 loci.
Materials and methodsCohortIn 2007, when the II National Survey of Drug Resistance was conducted, 20 Brazilian cities with a population of above 25,000 inhabitants, including the capital city of Belo Horizonte, with more than 10 new TB cases were selected and randomly distributed. The mycobacteriological tests (culture, identification and sensitivity test) were performed at the Ezequiel Dias Foundation (FUNED), the State Reference Center for TB. In 2007, 471 positive cultures referent to new TB cases were detected, excluding contaminated cultures and Non Tuberculous Mycobacteria (NTM) (9.3% – 44/471), leaving a final sample of 427. The result was possible in 70.5% (301/427) for the RFLP-IS6110 and 47.7% (204/427) for the MIRU-VNTR. However, for the comparative study between the RFLP-IS61103 and the MIRU-VNTR,7 204 new TB cases were identified, given that results were found in both techniques. The molecular tests were performed in the Molecular Biology Laboratory of the Social Pharmacy Department of the School of Pharmacy at the Federal University of Minas Gerais (UFMG).
RFLP-IS6110To perform the RFLP-IS6110, 3μL of the PvuII enzyme was used in a 1:2 proportion (enzyme and buffer) to restrict the genomic DNA. In each gel, three controls of the M. tuberculosis strain (Mt 14.323) were used, distributed in the 1st, 9th, and 18th wells. The transference to the membrane was performed manually, which was placed in contact with the gel for 24h. After the transference, the membrane was submitted to a process of colorimetric revelation with a phosphate and Nitroblue tetrazolium solution and then photographed.
An recent infection was defined when the results from the RFLP-IS6110 were greater than 85% similarity, and clusters were only considered when 100% of similarity was identified.11,12 The addresses of the patients were recorded to verify the epidemiological relationship (proximity of residence).
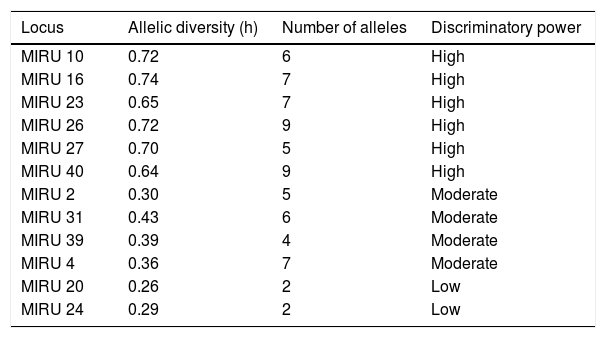
MIRU-VNTR 12 lociThe standardization was based on the 12 loci.7 To monitor and to guarantee quality and reproducibility, a negative sample and the DNA of the M. tuberculosis H37Rv, resulting from the PCR, were used as negative and positive internal controls, respectively. Ten samples were duplicated and implemented to control the reactions. The allelic diversity (h) that analyzes the variation of the different alleles found for the same locus within the samples was classified as: highly discriminant (h>0.6), moderately discriminant (0.3≤h≥0.6), and slightly discriminant (h<0.3).13
The recent infection was also defined for the MIRU-VNTR when the results were greater than 85% similarity, and clusters were only considered for strains with a 100% similarity.14,15
Statistical analysisVersion 7.1 of the BioNumerics software (AppliedMaths, Sint-Martens-Latem, Belgium) was used for the molecular analysis of the typing results from the RFLP-IS6110 and MIRU-VNTR, in turn building a dendrogram with all of the analyses for each technique in order to show the degree of similarity of the isolates by means of the Unweighted Pair Group Method of Arithmetic Average (UPGMA) and the Dice index with 1.5% of tolerance and 1.0% of optimization.3
Patterns of the RFLP-IS6110 were recorded with digitalized photographs, and the results of the typing from the MIRU-VNTR 12 loci were recorded using characters in a table.7
Statistical significance was measured to determine the proportion of confirmed or unconfirmed epidemiological relationships of isolates, which were grouped in RFLP-IS6110 and MIRU-VNTR in comparison with the subgroup of isolates grouped by RFLP-IS6110 but not by MIRU-VNTR. The analysis of logistic regression was performed to model the probability of finding an epidemiological relationship whether the isolate grouped by RFLP-IS6110 was confirmed or not by MIRU-VNTR, calculating the p-value according to the Mantel-Haenszel chi-square and Fischer Exact Odds Ratio (OR) with a Confidence Interval (CI) of 95%. The relationship between the epidemiological situation and the MIRU-VNTR within the same cluster was adjusted to the size of the cluster. All of the variables were analyzed by the OpenEpi software, version 3.0.
The discriminatory power described by Hunter and Gaston16 was used to determine if the molecular typing method was capable of distinguishing between two samples selected randomly from a given population. This calculation used the formula available at http://insilico.ehu.es/mini_tools/discriminatory_power/, which is based on the total number of isolates evaluated by a determined method of molecular typing. The value of D, which is the index of the discriminatory power, can vary from 0 to 1, where 0 means that all of the isolates of a specific population contain identical types and 1 indicates that the typing method was capable of distinguishing each isolate of a specific population from all others. The cluster rate was calculated according to the equation described by Bidovec-Stojkovic et al.,17 where T=(nc−c)/n, in which nc is the total number of grouped samples, c is the number of identified groups, and n is the total number of cases in the sample.
Grouping based on the RFLP-IS6110 and MIRU-VNTRTo compare the two methods of genotyping, the differences in the distribution of the clusters, the correlation with the results of the grouping, and the epidemiological relationship were analyzed.
The agreement in the grouping of cases between the two methods of typing was calculated by: (I) arrangement of fragmentation and identical profiles of RFLP-IS6110 and MIRU-VNTR; and (II) agreement in the labeling as a cluster or sole profile, regardless of the arrangement of the group.
To compare the standard deviation of the isolates, the groupings by RFLP-IS6110 were defined as the reference pattern.
EthicsThis study was approved by the ethics committee in research of the Federal University of Minas Gerais (ETIC 0416.0.203.000-10).
ResultsOf the 204 cultures that were submitted to the genotyping analysis through the RFLP-IS6110, a variation of 1–18 copies of IS6110 was observed, given that 107 (52.4%) presented 10–14 copies, 77 (37.7%) from 6 to 9 copies, 11 (5.3%) less than six copies, 7 (3.6%) from 15 to 17 copies, and 2 (1%) with 18 copies. Seven primary MDR-TB isolates were identified, all of which presented 8–15 copies of IS6110. The samples with less than six copies were not grouped in a cluster.
According to the dendrograms, 32% (66/204) of the isolates in the RFLP-IS6110 and 50.9% (104/204) of the isolates in the MIRU-VNTR had similarity equal to or greater than 85% (Fig. 1). A total of six (2.9%) of the isolates were included in the databank, indicated with an “empty space” and treated as a specific allele of the pattern (H37Rv positive control) due to non-amplification (Fig. 1).
Patterns are organized by similarity according to the dendrogram on the left. Bands were aligned adopting the MT 14323 strain as the reference, using the BioNumerics software, version 7.1 (Applied Maths, Sint-Martens-Latem, Belgium).
Table 1 describes the allelic diversity, the number of alleles, and the discriminatory power of MIRU-VNTR. It is important to note that the MIRU 20 and 24 showed a lower discriminatory power, with two alleles each (04/204), while the isolates of the MIRU-VNTR presented a similarity of equal to or greater than 85% (Fig. 1).
Allelic diversity of each locus and the discriminatory power.
| Locus | Allelic diversity (h) | Number of alleles | Discriminatory power |
|---|---|---|---|
| MIRU 10 | 0.72 | 6 | High |
| MIRU 16 | 0.74 | 7 | High |
| MIRU 23 | 0.65 | 7 | High |
| MIRU 26 | 0.72 | 9 | High |
| MIRU 27 | 0.70 | 5 | High |
| MIRU 40 | 0.64 | 9 | High |
| MIRU 2 | 0.30 | 5 | Moderate |
| MIRU 31 | 0.43 | 6 | Moderate |
| MIRU 39 | 0.39 | 4 | Moderate |
| MIRU 4 | 0.36 | 7 | Moderate |
| MIRU 20 | 0.26 | 2 | Low |
| MIRU 24 | 0.29 | 2 | Low |
Discriminatory power: high (h>0.6), moderate (0.3≤h≤0.6), and high (h<0.3).
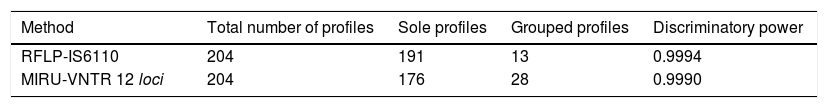
In the total of 204 genotypes, 191 sole profiles were found for RFLP-IS6110 and 176 for MIRU-VNTR. Of these, 6.4% (13/204) pertained to five clusters from RFLP-IS6110, while 13.7% (28/204) pertained to twelve clusters from the MIRU-VNTR. The agreement between the two genotyping techniques was of 87.7% (179/204), consisting of eight isolates in clusters and 171 isolates with a sole profile. The Kappa coefficient was 0.76 (good agreement). Of the 25 remaining isolates, 2.5% (5/204) were grouped by RFLP-IS6110 and 9.8% (20/204) by MIRU-VNTR. Three clusters were in agreement in both techniques.
Table 2 describes the total of sole and grouped profiles, as well as the discriminatory power from RFLP-IS6110 and MIRU-VNTR. The profiles grouped by RFLP-IS6110 were 6.4% (13/204) as compared to 13.7% (28/204) by MIRU-VNTR.
The cluster rate of RFLP-IS6110 and MIRU-VNTR was of 3.2% and 4.3%, respectively, and no significant difference could be observed (p>0.005).
Arrangement of the clusters by RFLP-IS6110 and MIRU-VNTRThe five clusters from RFLP-IS6110 consisted of 13 isolates, while the 12 clusters from MIRU-VNTR consisted of 28 isolates. The epidemiological relationship was significant for the RFLP clusters (4/13 patients – 30.7%) when compared with those from MIRU (4/28 patients – 14.2%) (p=0.002) (Fig. 2).
DiscussionThis is the first study in MG of M. tuberculosis genotyping that employs the RFLP-IS6110 and MIRU-VNTR. The high discriminatory power and the agreement demonstrate that both techniques can be used. However, the epidemiological relationship was greater when the clusters were detected by RFLP-IS6110 as compared to MIRU-VNTR (p=0.002); therefore, the RFLP proved to be more appropriate to evaluate this relationship when the clusters of M. tuberculosis were detected in MG.
The high variability of the number of copies detected by the RFLP-IS6110 of M. tuberculosis, as well as the genetic stability, make this technique a useful instrument for epidemiological diagnoses,10 although, when the genetic patterns are identical and with a low number of copies of IS6110, it may be deemed inappropriate. This low number of copies is one of the main factors that limits the broad application of this technique in Asian countries, such as India, China, and Thailand.17–22 Nevertheless, in the present study, this factor proved to be irrelevant, due to the low percentage (5.3%) of strains with less than six copies. Similar results were reported in the states of São Paulo, Goiás, Rio de Janeiro, and Rio Grande do Sul.15,23–25
The largest proportion of M. tuberculosis strains showed no recent transmission by IS6110 RFLP, used as the standard method, may well be due to the period of sample collection (only one year) and the type of selected individuals (new TB cases). Studies conducted in Brazil that report recent transmission (a similarity index>85%) included samples from a number of years.15,23,26 As regards the MIRU-VNTR, the present study found no difference between recent and late infections (50.9% index of similarity), especially since no other loci of MIRU-VNTR other than Mtub04, Mtub30, Mtub39, Mtub21, Qub4156, Qub11b, Qub26, ETRC, and ETRA were used.13
As regards the MIRU-VNTR in the use of molecular epidemiology, there was no need to use other loci, given that in this study the MIRU-VNTR 20 and 24 did not contribute to the differentiation of the strains, as described by other researchers.17,24,27
The high discriminatory power and agreement of both the RFLP-IS6110 and MIRU-VNTR demonstrates that they can be used.
Studies that show a high discriminatory power and agreement when comparing the RFLP-IS6110 and MIRU-VNTR were conducted by Allix-Beguec et al.28 in Belgium as well as in other countries, including Germany, Slovenia, and Brazil17,29,30 and showed results that were similar to the present study's results.
The limitation of this study lies in not having performed the MIRU15 and other loci13,28 that could detect the clusters with a greater epidemiological relationship as well as demonstrate the difference between recent and late infections. Studies that compared the two techniques and used the MIRU15 to identify the clusters presented more reliable results.17 Supply et al. proposed the 15-locus for epidemiological studies and 24-locus for phylogenetic studies typing of M. tuberculosis, because these methods can improve researches in the field of TB molecular epidemiology.7
Another limitation is that, Whole-genome sequencing (WSG) was not performed which could be an important molecular epidemiologic tool to provide additional resolution to strains with matching IS6110 RFLP, spoligotype or MIRU-VNTR loci profiles to more accurately distinguish patients who are part of a recent chain of transmission from those with reactivation disease related to distant infection. It is possible that WGS may become the standard method for strain typing for molecular epidemiological studies though efforts to address several limitations like high cost, the need for specialized software for data analysis, and incomplete understanding of the molecular clocks of SNPs (Single Nucleotide Polymorphisms) and LSPs (Large Nucleotide Polymorphisms). Further, the potential impact of WGS on patient care and design of public health strategies must be determined.31
This study was essential to gaining knowledge on molecular epidemiology in the state of MG. Although the molecular techniques of genotyping have evolved greatly over the years, each genetic marker unveils only a small portion of the information regarding genomics. Thus, the use of more than one technique when studying molecular epidemiology is warranted.
ConclusionThe RFLP-IS6110 and MIRU-VNTR proved to contain a high discriminatory power. The similarity index resulting from the RFLP showed no recent transmission. Good agreement was observed between the techniques when clusters were detected; however, the best epidemiological relationship was found when using the RFLP-IS6110.
Conflicts of interestThe authors declare no conflicts of interest.
The authors would like to thank all of the professionals who participated in this study, whether directly and indirectly, through the application of questionnaires. A special thanks goes out to staff members Carla and Ana Lethicia for their support in the Molecular Biology Laboratory, as well as to staff members Elza, Adriana, Wesley, Glaydson, and Alan for their work in the development of microbiological activities. This study was financially supported by the Foundation for Research Support of Minas Gerais (FAPEMIG – Process number – APQ 03266-13/APQ 00094-12), Coordination for the Improvement of Higher Education Personnel (CAPES), and the Ezequiel Dias Foundation, Minas Gerais, Brazil (FUNED).