β-Defensin-1, an antimicrobial peptide encoded by the DEFB1 gene, is known to play an important role in lung mucosal immunity. In our association study we analyzed three DEFB1 functional polymorphisms −52G>A (rs1799946), −44C>G (rs1800972) and −20G>A (rs11362) in 92 tuberculosis patients and 286 healthy controls, both from Northeast Brazil: no association was found between the studied DEFB1 polymorphisms and the disease. However we cannot exclude that this lack of association could be due to the low number of subjects analyzed, as suggested by the low statistical power achieved for the three analyzed SNPs (values between 0.16 and 0.50).
Mycobacterial diseases are a major health concern, affecting approximately one-third of the world's population.1 The most common clinical form of the disease is pulmonary tuberculosis, a granulomatous disease of the lungs caused by Mycobacterium tuberculosis. Extrapulmonary tuberculosis may occur when the bacillus infects other organs.2–4
The general risk of infected individuals to develop the disease during their lifetime ranges between 5 and 10%.2,3 A possible genetic predisposition to tuberculosis (TB) has been suggested in several studies.5–8 Moreover evidence exist that extrapulmonary TB is the result of an underlying immune defect,4 however, the functional correlation between genetic polymorphisms and immunologic defect should be yet unraveled.7
The mucosal immunity of air epithelia is complex, and the innate immune system, mainly by the mucosal secretion of antimicrobial peptides gradient, is thought to play an important role in defense against M. tuberculosis, although the exact mechanisms are yet poorly understood.9,10
The human β-defensin-1 (hBD-1) is a small (29–47 amino acids) cationic antimicrobial peptide,11 constitutively expressed in airway epithelia that plays an important role in mucosal immunity in the lung.12–15 This peptide is encoded by DEFB1 gene, located at chromosome 8p22-23.16
Three single nucleotide polymorphisms (SNPs), located at −52G>A (rs1799946), −44C>G (rs1800972) and −20G>A (rs11362) position in the 5′ untranslated region (UTR) of the DEFB1 gene have been reported to promote functional alteration of hBD-1 expression in different cell models.17,18 These SNPs were firstly described by Dörk and Stuhrmann19 and have been then associated with chronic obstructive pulmonary disease,20 increased levels of Candida albicans in the oral cavity,21 increased risk for Human Immunodeficiency Virus 1 infection,22–25 susceptibility to mycobacterial diseases8,26 and other diseases.27–31
Considering the possible role of the innate immune system in the risk of tuberculosis infection, this study aims to examine whether functional polymorphisms at 5′UTR in DEFB1 gene are associated with the occurrence of active tuberculosis.
Patients and methodsStudy populationNinety-two TB patients were enrolled from five reference center for TB follow up in Recife (Pernambuco, Northeast Brazil): Hospital das Clínicas – Universidade Federal de Pernambuco (HC-UFPE), Instituto de Medicina Integral Professor Fernando Figueira (IMIP), Hospital Barão de Lucena (HBL/US) and Hospital Otávio de Freitas. Of the 92 patients analyzed, 64 (69.6%) presented with pulmonary TB (36 males and 28 females, mean age 21 years±18.7) and 28 (30.4%) with extrapulmonary TB (17 males and 11 females, mean age 23 years±21.7). The diagnosis was based on clinical symptoms and radiographic findings, along with bacteriological confirmation (culture, smear and/or polymerase chain reaction) as described by the Diagnostic Standards and Classification of Tuberculosis in Adults and Children.32
As control group, we enrolled 286 healthy individuals (163 females/123 males; mean age 27 years±20.3), unrelated to patients, with negative Mantoux test, showing no symptoms of tuberculosis or previous history of the disease. All patients and control subjects came from the same geographical area (Pernambuco, Brazil), were all HIV-1 negative and were not under immunosuppressive medication.
Since ethnicity is an important issue in genetic analyses, patients and controls reported their self-declared skin based ethnic origin. During ethnicity report a health professional accompanied the procedure guaranteeing the correctness of the procedure. After ethnicity assessment we verified the self-reported and health professional checked results, by analyzing in the controls 12 Ancestry informative markers (AIMs) as recently reported by Coelho et al.33 Absence or extremely poor correlation between skin based ethnicity and the true genetic background, as revealed by the 12 AIMs analysis, was observed. So, we discarded the self-declared skin based ethnic origin and considered only the genetic AIMs findings. In fact, our studied population consists of a strong admixture of European-Caucasian, African and South American Amerindians population, leading the genetic North East Brazilian composition unique; consequently it is impossible to stratify our population for ethnicity, being an admixture of around 58% European, 24% African and 18% Amerindian as recently reported.33
Written informed consent was obtained from the participants or their parents. The CPqAM/FIOCRUZ Ethics Committee (CEP Registration – 55/05) approved the study. Patients underwent a standardized clinical–epidemiological questionnaire.
DEFB1 genotypingGenomic DNA was extracted from whole blood using QIAamp DNA Blood Kit according to manufacturer instructions (QIAamp DNA Blood Midi Kit, Qiagen).
The DEFB1 5′UTR polymorphisms −52G>A (rs1799946), −44C>G (rs1800972) and −20G>A (rs11362) were genotyped by direct sequencing, according to Braida et al.22 Sequences were handled using the 4Peaks (http://nucleobytes.com/index.php/4peaks) and CodonCode Aligner software (http://www.codoncode.com/aligner/).
Statistical analysisChi-square test was used to verify the Hardy–Weinberg equilibrium and the Fisher's exact test was employed for pair-wise comparison of allele, genotype and haplotype frequencies using contingency tables as appropriate; only p values<0.05 were considered to be significant. All the statistical analyses were carried out using the open-source R package (http://www.r-project.org). The power of the study was checked by software G*power software version 3.1.34 Haplotypes were computed using the Arlequin version 3.5.1.335 and Haploview software.36
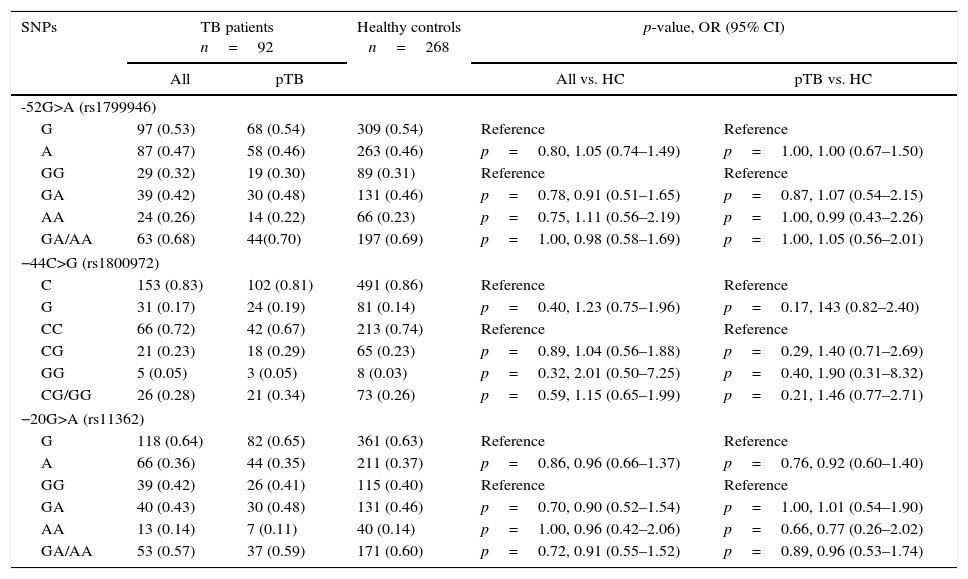
ResultsThe genotyping results of the three 5′UTR DEFB1 functional polymorphisms −52G>A (rs1799946), −44C>G (rs1800972) and −20G>A (rs11362) in TB patients and healthy controls, are shown in Table 1. All SNPs were in Hardy–Weinberg equilibrium for both TB patients and healthy controls.
Alleles and genotypes distribution of SNPs – in the 5′ untranslated region (UTR) of DEFB1 gene in tuberculosis patients (TB) and healthy controls (HC) from Northeast Brazil.
| SNPs | TB patients n=92 | Healthy controls n=268 | p-value, OR (95% CI) | ||
|---|---|---|---|---|---|
| All | pTB | All vs. HC | pTB vs. HC | ||
| -52G>A (rs1799946) | |||||
| G | 97 (0.53) | 68 (0.54) | 309 (0.54) | Reference | Reference |
| A | 87 (0.47) | 58 (0.46) | 263 (0.46) | p=0.80, 1.05 (0.74–1.49) | p=1.00, 1.00 (0.67–1.50) |
| GG | 29 (0.32) | 19 (0.30) | 89 (0.31) | Reference | Reference |
| GA | 39 (0.42) | 30 (0.48) | 131 (0.46) | p=0.78, 0.91 (0.51–1.65) | p=0.87, 1.07 (0.54–2.15) |
| AA | 24 (0.26) | 14 (0.22) | 66 (0.23) | p=0.75, 1.11 (0.56–2.19) | p=1.00, 0.99 (0.43–2.26) |
| GA/AA | 63 (0.68) | 44(0.70) | 197 (0.69) | p=1.00, 0.98 (0.58–1.69) | p=1.00, 1.05 (0.56–2.01) |
| −44C>G (rs1800972) | |||||
| C | 153 (0.83) | 102 (0.81) | 491 (0.86) | Reference | Reference |
| G | 31 (0.17) | 24 (0.19) | 81 (0.14) | p=0.40, 1.23 (0.75–1.96) | p=0.17, 143 (0.82–2.40) |
| CC | 66 (0.72) | 42 (0.67) | 213 (0.74) | Reference | Reference |
| CG | 21 (0.23) | 18 (0.29) | 65 (0.23) | p=0.89, 1.04 (0.56–1.88) | p=0.29, 1.40 (0.71–2.69) |
| GG | 5 (0.05) | 3 (0.05) | 8 (0.03) | p=0.32, 2.01 (0.50–7.25) | p=0.40, 1.90 (0.31–8.32) |
| CG/GG | 26 (0.28) | 21 (0.34) | 73 (0.26) | p=0.59, 1.15 (0.65–1.99) | p=0.21, 1.46 (0.77–2.71) |
| −20G>A (rs11362) | |||||
| G | 118 (0.64) | 82 (0.65) | 361 (0.63) | Reference | Reference |
| A | 66 (0.36) | 44 (0.35) | 211 (0.37) | p=0.86, 0.96 (0.66–1.37) | p=0.76, 0.92 (0.60–1.40) |
| GG | 39 (0.42) | 26 (0.41) | 115 (0.40) | Reference | Reference |
| GA | 40 (0.43) | 30 (0.48) | 131 (0.46) | p=0.70, 0.90 (0.52–1.54) | p=1.00, 1.01 (0.54–1.90) |
| AA | 13 (0.14) | 7 (0.11) | 40 (0.14) | p=1.00, 0.96 (0.42–2.06) | p=0.66, 0.77 (0.26–2.02) |
| GA/AA | 53 (0.57) | 37 (0.59) | 171 (0.60) | p=0.72, 0.91 (0.55–1.52) | p=0.89, 0.96 (0.53–1.74) |
pTB, pulmonary tuberculosis; OR, odds ratio; 95% CI, 95% confidence interval; SNP, single nucleotide polymorphism. Reference indicates the most frequent alleles and genotypes.
DEFB1 SNPs allele and genotype frequencies were similar in TB patients and healthy controls and no significant difference has been found (p>0.05), also when TB patients were stratified according to the pulmonary form of disease.
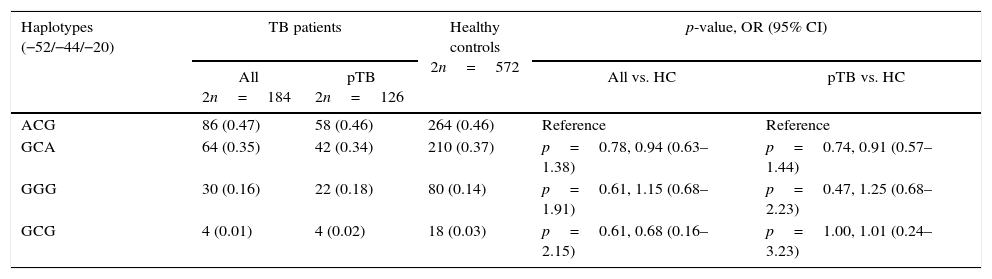
We computed the haplotypes and observed that the three SNPs in the 5′UTR of DEFB1 are in strong linkage disequilibrium (D′>0.9), forming four haplotypes (Table 2). However, no significant differences were found when comparing TB patients and healthy controls (Table 2).
Haplotypes frequency distribution of 5′UTR DEFB SNPs in tuberculosis patients (TB) and healthy controls (HC) from Northeast Brazil.
| Haplotypes (−52/−44/−20) | TB patients | Healthy controls 2n=572 | p-value, OR (95% CI) | ||
|---|---|---|---|---|---|
| All 2n=184 | pTB 2n=126 | All vs. HC | pTB vs. HC | ||
| ACG | 86 (0.47) | 58 (0.46) | 264 (0.46) | Reference | Reference |
| GCA | 64 (0.35) | 42 (0.34) | 210 (0.37) | p=0.78, 0.94 (0.63–1.38) | p=0.74, 0.91 (0.57–1.44) |
| GGG | 30 (0.16) | 22 (0.18) | 80 (0.14) | p=0.61, 1.15 (0.68–1.91) | p=0.47, 1.25 (0.68–2.23) |
| GCG | 4 (0.01) | 4 (0.02) | 18 (0.03) | p=0.61, 0.68 (0.16–2.15) | p=1.00, 1.01 (0.24–3.23) |
pTB, pulmonary tuberculosis; OR, odds ratio; 95% CI, 95% confidence interval; nc, not calculable. Reference indicates the most frequent haplotypes.
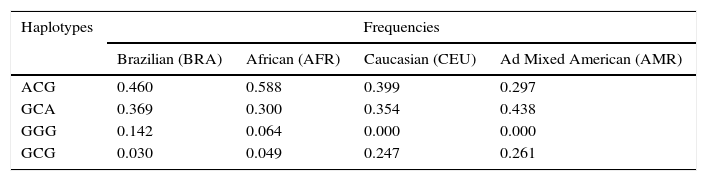
In addition, we estimated the linkage disequilibrium and the possible haplotypes, using Haploview software, for Brazilian healthy controls (BRA) and healthy individuals of African (AFR), Caucasian (CEU) and Ad Mixed American (AMR) origin, provided by 1000 Genome Project Database (www.1000genomes.org). We noted that DEFB-1 SNPs were in perfect linkage disequilibrium (D′=1.0) and have different haplotype distributions in the studied populations (Table 3).
The post hoc analysis of the study through the G*power software, indicate the following power for three SNPs: 0.30 (−52G>A), 0.5 (−44C>G) and 0.16 (−20G>A).
DiscussionIn the current study, no association was found between −52G>A (rs1799946), −44C>G (rs1800972), −20G>A (rs11362) DEFB1 polymorphisms and TB in a Brazilian population, also when considering the pulmonary form of the disease.
In a case–control study, Wu et al.8 analyzed DEFB1 5′UTR polymorphisms in 102 patients with pulmonary TB and 148 healthy controls of Chinese Han population observing that the DEFB1 −44C/C genotype was associated with pulmonary TB susceptibility (OR=2.096) and the GGG (−52/−44/−20) haplotype was associated with the pulmonary TB protection (OR=0.348).
In a recent work, performed on a limited number of Mexican patients (27 with pulmonary TB and 16 with extrapulmonary TB) and controls (n=49), López Campos et al.37 described an association between DEFB1 −44C>G SNP (CG genotype in a codominant genetic model) and susceptibility to extrapulmonary TB, but not with the pulmonary TB, hypothesizing a role for hBD-1 in the fight against M. tuberculosis outside the lung.
Our findings have discrepancies with the results obtained by Wu et al.8 on Han population and by Lopez Campos et al.37 in Mexicans: we can hypothesize that the differences could be, at least in part, due to ethnicity, since the frequencies DEFB1 SNPs vary in different populations. According to the NCBI and HapMap databases,38 for −52 SNP (rs1799946) the frequency of the ancestral allele is 0.64 in European (CEU), 0.56 in Sub-Sahara African (YRI) and 0.40 in Asian (HCB); the frequency of ancestral allele of −44 SNP (rs1800972) is 0.26 in European (CEU), 0.04 in Sub-Sahara African (YRI) and 0.12 in Asian (HCB); for −20 SNP (rs11362), the frequency is 0.61 in European (CEU), 0.60 in Sub-Sahara (YRI) and 0.71 in Asian (HCB). In addition, the SNPs analyzed in Brazilian healthy individuals and healthy individual of African (AFR), in Caucasian (CEU) and Ad Mixed American (AMR) origin, were in perfect linkage disequilibrium (D′=1.0) and presented different haplotypic distribution (Table 3), suggesting that ethnics background may be responsible for the differences observed in populations that have been studied. Additionally, in the Han Chinese population no linkage disequilibrium was detected (D′<0.5)8 and as a consequence comparing DEFB1 haplotype results with our findings has not been possible. Moreover, when looking at the Mexican population it has to be remarked that the study of Lopez Campos et al.37 just analyzed one DEFB1 SNP −44C>G and other SNPs in cathelicidin encoding CAMP gene. So, it was not possible to compare our DEFB1 haplotypes with the results reported for Mexican patients. Furthermore, DEFB1 rs1800972 allele and genotype frequencies reported in the Mexican population (not in Hardy–Weimberg Equilibrium) were significantly different (0.0002) for the C allele to Brazilian ones.
However, we cannot exclude, that the lack of association reported in our study, could be due to the limited sample size; the power of our study (post hoc Power≤0.5) indicates that not finding an association between SNPs with TB development, could be due to a type II error. This is a strong limitation of our study, and seeing that also the works of Wu et al.8 and Lopez Campos et al.37 suffer the same sample size problem, we are convinced that any further study dealing with association between defensins gene polymorphism and TB, should enroll greater number of patients, may be joining several research groups, in order to achieve better statistical power to provide a definitive answer on the role of defensins in susceptibility to TB.
All this considered, even if our results seem to exclude an association between DEFB1 polymorphisms and TB development in Northeast Brazilian population, since DEFB1 could be a potential candidate gene for susceptibility to TB and only very few studies have addressed this topic achieving controversial results, we do think that further studies on a larger number of patients and in other populations are needed to disclose the role of DEFB1 variations in tuberculosis development.
Conflicts of interestThe authors declare no conflicts of interest.
The authors thank all the health professionals from the hospitals involved in the study that collaborated with the research. We thank the Laboratory of Immunoepidemiology – Centro de Pesquisas Aggeu Magalhões/Fundação Oswaldo Cruz (CPqAM/FIOCRUZ), the Laboratory of Immunopathology Keizo Asami, the Department of Genetics, Federal University of Pernambuco, the Graduate Program in Genetics for supporting physical and scientific, as well as Fundação de Amparo à Ciência e Tecnologia do Estado de Pernambuco (FACEPE), Coordenação de Aperfeiçoamento Pessoal de Nível Superior (CAPES) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), for financial support.