Mamastrovirus 5 (MAstV5), belonging to the Astroviridae (AstV) family, previously known as canine astrovirus or astrovirus-like particles, has been reported in several countries to be associated with viral enteric disease in dogs since the 1980s. Astroviruses have been detected in fecal samples from a wide variety of mammals and birds that are associated with gastroenteritis and extra enteric manifestations. In the present study, RT-PCR was used to investigate the presence of MAstV5 in 269 dog fecal samples. MAstV5 was detected in 26% (71/269) of the samples. Interestingly, all MAstV5-positive samples derived from dogs displaying clinical signs suggestive of gastroenteritis, other enteric viruses were simultaneously detected (canine parvovirus, canine distemper virus, canine coronavirus, canine adenovirus and canine rotavirus). Based on genomic sequence analysis of MAstV5 a novel classification of the species into four genotypes, MAstV5a-MAstV5d, is proposed. Phylogenetic analyses based on the ORF2 amino acid sequences, samples described herein grouped into the putative genotype ‘a’ closed related with Chinese samples. Other studies are required to attempt the clinical and antigenic implications of these astrovirus genotypes in dogs.
Viruses belonging to the Astroviridae (AstV) family are spherical, non-enveloped, 28–30nm in size, with a surface that forms a characteristic star-like structure.1 The RNA genome of AstV ranges from 6.8 to 7.9-kb in size, polyadenylated at the 3′ end, and contains three ORFs designated as ORF1a, ORF1b and ORF2. ORF1a encodes a protease, ORF1b encodes an RNA-dependent RNA-polymerase,2,3 while ORF2 encodes the viral capsid structural polyprotein that is required for virion assembly.4
The viral classification was previously based on the host and consisted of two genera, Avastrovirus and Mamastrovirus. However, recent characterization of novel astroviruses has taken in consideration that isolates from different animal species can be genetically similar, while genetically diverse viruses can be isolated from the same animal species.2 Based on this analysis, the International Committee on Taxonomy of Viruses renamed canine astrovirus as Mamastrovirus 5 (MAstV5).5
Astroviruses have been detected in fecal samples from a wide variety of mammals and birds that are associated with gastroenteritis.2 In children, AstVs are the second most common cause of gastroenteritis after rotaviruses.2,6 Human AstVs can also cause significant disease in the elderly7 and immune-compromised patients.8,9 In addition to enteric manifestations, AstVs have been associated with fatal hepatitis in ducks,10 interstitial nephritis in young chickens,11 stunting and pre hatching mortality in duck and goose embryos,12 as well as shaking mink syndrome13. Recently, an AstV was also hypothesized to be the causative agent of nonsuppurative encephalitis in cattle.14
Since the 1980s, astrovirus-like particles have been reported in dogs with and without diarrhea.15–17 To date, canine astroviruses or astrovirus-like particles infecting dogs have been reported in several countries.15–25 Despite the detection of MAstV5 in association with gastroenteritis in dogs, which suggests a possible role for MAstV5 as a canine enteric pathogen, the association of MAstV5 with clinical disease remains obscure in such reports. Here, we investigated the presence of MAstV5 using RT-PCR in fecal samples from dogs of different ages with and without diarrhea. The partial genomes of selected MAstV5 RNA-positive samples were also sequenced to perform a phylogenetic analysis comparing them with the MAstV5 sequences described in the literature as the cause of enteric disease.18,20,23,26 Additionally, MAstV5 was proposed to be classified in four putative genotypes.
Materials and methodsSamples and nucleic acid extractionA total of 269 dog fecal samples were collected between 2008 and 2014 in veterinary clinics and hospitals by convenience. These samples were obtained from eight Federal States of Brazil (Acre, Mato Grosso do Sul, Paraná, Rio Grande do Sul, Rio de Janeiro, Rondônia, Santa Catarina and São Paulo). The animal's age was recorded and ranked from puppy (equal or less than one-year-old) to adult dog (more than one-year-old); some samples from dogs of unknown age were included. Animals not presenting diarrhea at the time of sampling were considered asymptomatic and those presenting clinical signs of enteric disease diarrhea were classified as symptomatic. Samples were diluted to 20% (w/v) in phosphate buffered saline (PBS, pH 7.4) and stored at −80°C for further analysis. Subsequently, viral DNA isolation from the supernatant was performed using a commercial kit (NewGene Preamp®, Simbios Biotecnologia, Brazil) based on guanidine isothiocyanate and silica.27 Viral RNA was isolated using TRIzol® LS Reagent (Life Technologies™, USA) according to the manufacturer's instructions.
Oligonucleotides for MAstV5 detection and sequencingAn initial screening using RT-PCR to detect a larger number of Mamastrovirus species was achieved by amplifying 422bp of the ORF1b fragment using oligonucleotides, as previously described.28 For the specific detection of MAstV5, 92 nucleotide sequences of this species were retrieved from GenBank database (http://www.ncbi.nlm.nih.gov/nucleotide), and aligned using CLUSTAL W within Molecular Evolutionary Genetics Analysis version 6 (MEGA6).29 The MAstV5 specific RT-PCR was designed with a primer pair targeting the region of ORF2 that amplified a 250bp fragment selected using Primer3 software.30 In addition, the 16S rRNA gene from Escherichia coli was amplified using the primer pair FC27 and R530 as an endogenous internal control in each fecal sample evaluated for the specific presence of MAstV5.31 For partial genome amplification, sets of 12 pairs of sequencing primers were selected to amplify overlapping fragments of ORF1 (ORF1a and ORF1b) and capsid protein (ORF2) segment representing a consensus sequence of approximately 5000 nucleotides. The primer sequences are shown in Table 1.
Oligonucleotide sequences applied in the present work.
| Primer name | Sequence (5′–3′) | Target | Objective | Reference |
|---|---|---|---|---|
| ADS_138F | AATGTCACGGGGATACCATC | ORF1a | Phylogenetic analysis | Present study |
| ADS_230F | GTGCATCAAACCAACACTGG | |||
| ADS_690R | GGGTCACTCCATTCAGGAAA | |||
| ADS_367F | CAAACCCCAACCTCAAGAGA | |||
| ADS_941R | CATCCTCAGCAGTCCAGTCA | |||
| ADS_719F | TGGGACACATATGGTGATGAA | |||
| ADS_1022R | GCCTAGGCTTGAGGATGTGA | |||
| ADS_922F | TGACTGGACTGCTGAGGATG | |||
| ADS_1606R | AGTCGGCTTCGGTGTCATAG | ORF1b | ||
| ADS_1317F | TGATCCGCTCAAATCCCTAC | |||
| ADS_1669R | TTGCTCCGGACATAATCCTC | |||
| ADS_1565F | CTGCCTATCCCAAGATGCTC | |||
| ADS_2192R | GCAAATTCAAAAGCCTGGAG | |||
| ADS_2173F | CTCCAGGCTTTTGAATTTGC | |||
| ADS_2857R | ACTCTCTTGCGACCACGATT | ORF2 | ||
| ADS_2839F | ATCGTGGTCGCAAGAGAGTT | |||
| ADS_3535R | AGTGGTTGTCCTGCTTCACC | |||
| ADS_3415F | TTGAGCTTCACTGCACTTGG | |||
| ADS_4033R | CATGGTGGGTTCTGTTGGTA | |||
| ADS_3963F | CCAGCTGTTATTGGGGACAA | |||
| ADS_4628R | TTGGTGGTGTTCTGAGGAAA | |||
| ADS_4446F | GCCCCTGGTTCATTTTTGT | |||
| ADS_5030R | TGAACCTGTACCCTCGATCC | |||
| ADS_TTTR | TTTTTTTTTTTTTTTTTTTT | Poli A tail | ||
| ASTRO 2 | GARTTYGATTGGRCKCGKTAYGA | ORF 1b | Screening | 28 |
| ASTRO 3 | GGYTTKACCCACATNCCRAA | |||
| ASTRO 4 | CGKTAYGATGGKACKATHCC | |||
| ASTRO 5 | AGGTAYGATGGKACKATHCC | |||
| ASTRO 6 | GARTTYGATTGGRCKAGGTAYGA | |||
| AstVCan For | TCTGATGATGATTCTCTTCTTGATG | ORF2 | MAstV5 specific | Present study |
| AstVCan Rev | GGGAACACTTTTCACGAGCA | |||
| FC27 | AGAGTTTGATCCTGGCTCAG | 16S RNA E. coli | Internal control | 31 |
| R530 | CCGCGGCTGCTGGCACGTA | |||
| CPV 555 F | CAGGAAGATATCCAGAAG | VP2 | CPV detection | 35 |
| CPV 555 R | GGTGCTAGTTGATATGTA | |||
| HA1 | CGCGCTGAACATTACTACCTTGTC | E3 | CAdV detection | 36 |
| HA2 | CCTAGAGCACTTCGTGTCCGCTT | |||
| CCoV 1F | TCCAGATATGTAATGTTCGG | M | CCoV detection | 37 |
| CCoV 2R | TCTGTTGAGTAATCACCAGCT | |||
| BEG 9F | GGCTTTAAAAGAGAGAATTTCCGTCTGG | VP7 | CRV detection | 38 |
| END 9R | GGTCACATCATACAATTCTAATCTAAG | |||
| CDV 1F | ACTGCTCCTGATACTGC | NC | CDV detection | 39 |
| CDV 2R | TTCAACACCRACYCCC | |||
| CDV 3F | ACAGRATTGCYGAGGACYTRT | 40 | ||
| CDV 4F | CARRATAACCATGTAYGGTGC | |||
VP, virus protein; ORF, open read frame; E, early region; M, structural protein M; NC, nucleocapsid protein.
The cDNA was synthesized using SuperScript® III Reverse Transcriptase Kit (Life Technologies, USA) using the reverse primers in a total volume of 20μL, following the manufacturer's instructions. The cDNA amplification was conducted in a final volume of 25μL containing 1× PCR buffer, 1.5mM of MgCl2, 0.2mM of dNTP mix, 0.2μM of each primer and 1 unit of Platinum® Taq DNA Polymerase (Life Technologies, USA). The first round of RT-PCR screening was carried out with an initial incubation at 94°C for 3min, 30 cycles of amplification consisting of denaturation at 94°C for 1min, annealing at 50°C for 1min, and extension at 72°C for 1min. The second round was performed in a final volume of 25μL that contained 2μL of the first reaction product and the thermocycler conditions were the same as those used for the first round.
The MAstV5-specific RT-PCR with specific and internal control primers was performed as a multiplex protocol. Cycling conditions were an initial cycle at 94°C for 5min, 25 cycles of denaturation at 94°C for 30s, annealing at 58°C for 30s and polymerization at 72°C for 1min, which was followed by a final extension cycle at 72°C for 7min. To confirm the specific amplification of MAstV5, RT-PCR products were submitted to purification using the NucleoSpin Extract II Kit (Macherey-Nagel, Germany) and sequenced. Both DNA strands were sequenced with an ABI PRISM 3100 Genetic Analyzer using a BigDye Terminator v.3.1 cycle Sequencing Kit (Applied Biosystems, USA).
Detection of other enteric virusesAll positive MAstV5 samples were also screened for other common enteric viruses through the amplification of cDNA/DNA. The primer pairs used for the detection of canine distemper virus (CDV), carnivore protoparvovirus 1 (canine parvovirus 2, CPV2), canine coronavirus (CCoV), canine rotavirus (CRV) and, canine adenovirus 1 (CAdV1) and CAdV2 are shown in Table 1. The cDNA/DNA amplification of the target sequences was conducted in a total volume of 25μL containing 1× PCR buffer, 1.5mM of MgCl2, 0.2mM of dNTP mix, 0.2μM of each primer pair and 1 unit of Taq DNA Polymerase (Ludwig Biotecnologia, Alvorada, RS, Brazil).
Genome amplificationFour MAstV5-positive samples were selected, taking into account their different geographical origins. ORF1a, ORF1b and ORF2 sequences were amplified using a nested touchdown RT-PCR method. The first round of amplification was conducted in a final volume of 25μL. The cycling conditions included an initial denaturation at 95°C for 5min, 20 cycles of 30s for denaturation at 95°C, outer primer pair annealing for 30s at 55–45°C per the touchdown method, and 7min of extension at 72°C, with a final 7min extension at 72°C. The same final volume was used in the second round, which contained 2μL of the amplification product of the first round. The second round cycling conditions were an initial denaturation at 95°C for 5min, 30 cycles of 30s of denaturation at 95°C, inner primer pair annealing for 30s at 55–45°C per the touchdown method, and 1min of extension at 72°C, with a 7min final extension at 72°C.
Sequencing and phylogenetic inferencesThe RT-PCR products generated with the sets of sequencing primers were purified using the NucleoSpin Extract II Kit (Macherey-Nagel, Germany). Both DNA strands were sequenced with an ABI PRISM 3100 Genetic Analyzer using a BigDye Terminator v.3.1 cycle Sequencing Kit (Applied Biosystems, USA). Overlapping fragments were aligned and assembled using SeqMan software from the DNASTAR package (DNASTAR, USA).32 [The open reading frames were identified using the NCBI ORF Finder software (http://www.ncbi.nlm.nih.gov/gorf/gorf.html).]
Sequence alignment was performed using the CLUSTAL W. For the phylogenetic inferences of the MAstV capsid protein region, the MAstV5 sequences that were submitted to genome amplification and 12 MAstV5 representative strains were included. MEGA6 software29 was used for phylogeny inference calculated using “find best DNA/protein model” tool from MEGA6. The Kimura 2-parameter substitution model was selected for the MAstV5 ORF2 nucleotide inference, and the LG substitution model (frequencies +F) was used for the amino acid inference. The substitution-rate variation among sites was modeled with a gamma distribution (shape parameter=5). Statistical support was provided by 1000 non-parametric bootstrap analyses. A nucleotide distance matrix was calculated using an alignment with ORF2 and partial genome sequences (Table 2). The Mamastrovirus genotypes were distinguished based on the amino acid sequence of the full length ORF2, where the genetic distances (p-dist) 0.378–0.750 and 0.006–0.312 between and within groups, respectively, were used.5 All of the sequence alignments used to construct the phylogenetic trees are available in Figshare (http://figshare.com/) with the DOI number https://doi.org/10.6084/m9.figshare.4596325. The nucleotide sequences obtained in this study were deposited in GenBank under accession numbers KR349488–KR349491.
Comparison of identity percentage between nucleotide sequence of the partial genomes and amino acid sequence identity percentage of open reading frame 2 (ORF2) from the sequenced MAstV5 compared with sequences available in GenBank.
| Strain | GenBank accession no. (genome size) | Sara/13/BRA | 5617/12/BRA | GRAV/13/BRA | 237/13/BRA | ||||
|---|---|---|---|---|---|---|---|---|---|
| (4980 nt, KR349488) | (5012 nt, KR349490) | (5039 nt, KR349491) | (5011 nt, KR349489) | ||||||
| Partial genome | ORF2 | Partial genome | ORF2 | Partial genome | ORF2 | Partial genome | ORF2 | ||
| Bari/2008_ITA | HM045005 (3120 nt) | 82 | 84 | 81 | 84 | 82 | 84 | 81 | 84 |
| Italy/2005 | FM213330 (2756 nt) | 79 | 80 | 79 | 80 | 79 | 81 | 79 | 80 |
| GI.E/Dog/ITA/2010/Zoid | JN193534 (2949 nt) | 76 | 77 | 76 | 77 | 75 | 77 | 76 | 77 |
| China/2008_SH8 | HQ623147 (2738 nt) | 94 | 97 | 96 | 97 | 95 | 96 | 96 | 97 |
| China/2008_SH15 | HQ623148 (2738 nt) | 94 | 97 | 96 | 97 | 95 | 96 | 96 | 97 |
| Gillingham/2012/UK | NC_026814 (6617 nt) | 86 | 79 | 86 | 79 | 86 | 80 | 86 | 79 |
| Lincoln/2012/UK | KP404150 (6613 nt) | 95 | 97 | 95 | 97 | 94 | 95 | 96 | 97 |
| HUN/2012/2 | KX599349 (6587 nt) | 94 | 97 | 95 | 97 | 94 | 95 | 94 | 97 |
| HUN/2012/6 | KX599350 (6576 nt) | 87 | 80 | 87 | 81 | 87 | 81 | 87 | 80 |
| HUN/2012/115 | KX599351 (6569 nt) | 86 | 78 | 85 | 78 | 85 | 77 | 86 | 78 |
| HUN/2012/126 | KX599352 (6535 nt) | 76 | 76 | 76 | 76 | 75 | 75 | 76 | 76 |
| HUN/2012/135 | KX599353 (6571 nt) | 92 | 95 | 92 | 94 | 91 | 93 | 92 | 94 |
The RT-PCR protocol using the screening primers for MAstV5 was positive in 22% (64/269) of the samples, and the protocol using the specific primers for MAstV5 identified 12% (32/269) of the samples tested. The sample was considered MAstV5 positive if the results of at least one of the two RT-PCR protocols were positive, which resulted in 26% (71/269) of the fecal samples being detected as positive. In addition, PCR products generated from both protocols were submitted to DNA sequencing to confirm the results (data not shown). The RT-PCR internal control from the 16S rRNA gene of E. coli was positive in all 269 of the samples tested.
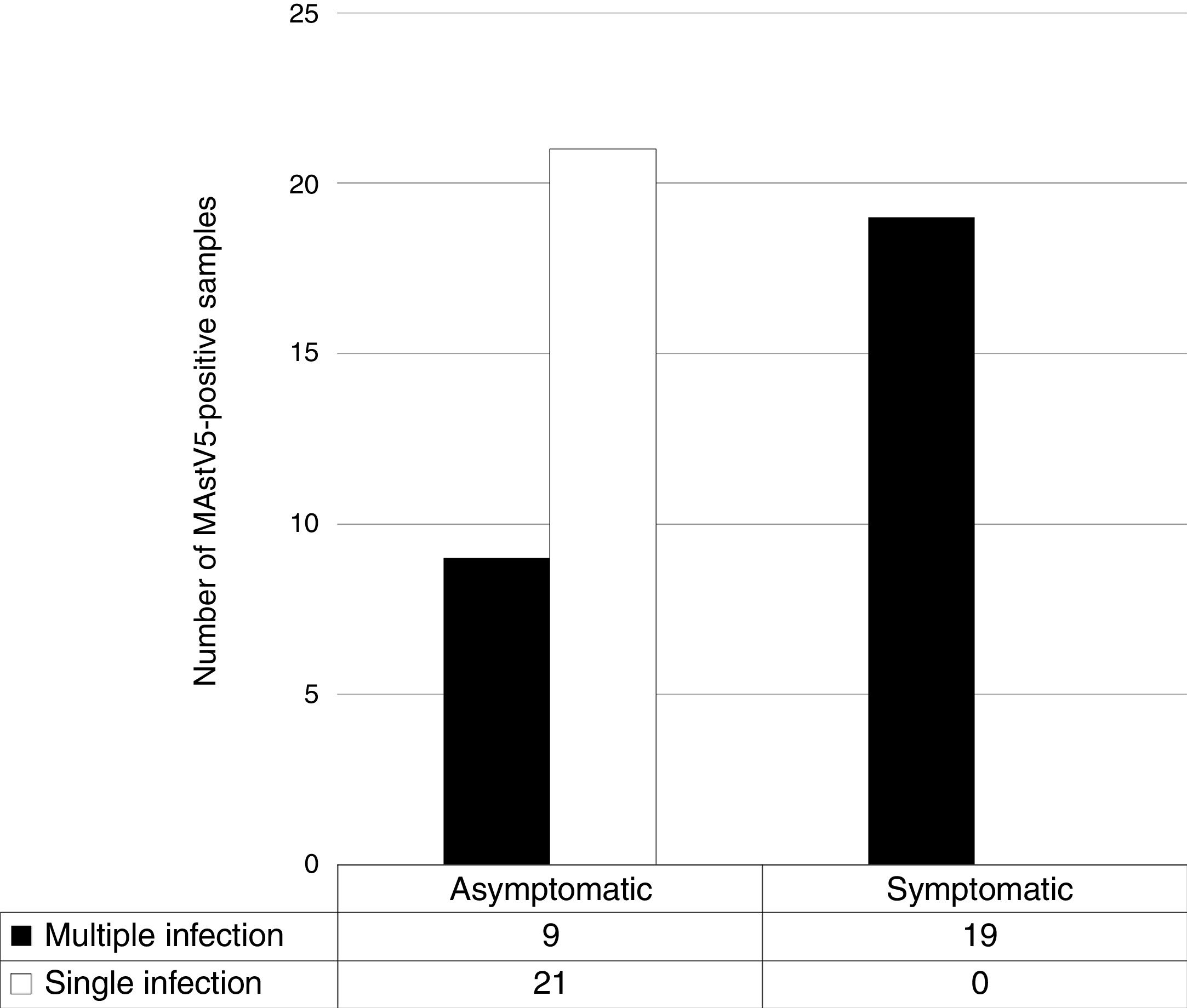
Information about clinical signs of enteric disease was available for 49 of the 71 positive samples. Considering only these, in all of the MAstV5-positive samples derived from dogs with clinical signs suggestive of gastroenteritis, other enteric viruses were simultaneously detected. Detailed results for the supposed association with clinical signs and the detection of other enteric viruses are shown in Fig. 1 and Table 3, respectively.
Detection of dog enteric viruses in the 49 MAstV5-positive samples.
| Virus | n | % |
|---|---|---|
| MAstV5 alone | 21 | 43 |
| CDV+MAstV5 | 9 | 18 |
| CPV+MAstV5 | 3 | 6 |
| CRV+MAstV5 | 1 | 2 |
| CCoV+MAstV5 | 4 | 8 |
| CDV+CPV+MAstV5 | 6 | 12 |
| CPV+CCoV+MAstV5 | 1 | 2 |
| CPV+CAV+MAstV5 | 1 | 2 |
| CDV+CPV+CCoV+MAstV5 | 1 | 2 |
| CPV+CCoV+CRV+MAstV5 | 1 | 2 |
| CDV+CPV+CCoV+CAV+MAstV5 | 1 | 1 |
| Total | 49 | 100 |
Animals were ranked in age category as puppies, adults and unknown age. Adults revealed a higher frequency of MAstV5-positive RT-PCR results than puppies (puppies: 44/166, 26.5%; adults: 6/16, 37.5%; unknown age: 21/91, 23.1%). No significant difference between dogs age and MAstV5 positive samples was observed in the different ranked ages (P=0.56).
MAstV5 genome sequences and phylogenetic inferencesFour partial genomes were obtained, namely, MAstV5_Sara/13/BRA, MAstV5_237/13/BRA, MAstV5_GRAV/13/BRA and MAstV5_5617/12/BRA, which were 4980, 5011, 5039 and 5012 nt in length, respectively, excluding the poly(A) tail and the 5′ untranslated regions (UTRs). The three first partial genomes were collected from different cities of Rio Grande do Sul State (Porto Alegre, Viamão and Gravataí, respectively), and one was from Londrina city, Paraná State. All of the four nearly complete genomes contained a typical AstV organization in the three predicted ORFs – ORF1a, ORF1b and ORF2. Further genome sequence comparison revealed that the four partial genomes had greater identities (94–96%) with Chinese strains, considered as the Italy, Hungary and United Kingdom strains identity ranged from 76–82%, 75–95% and 86–96% when compared to the Brazilian strains identified here, respectively (Table 2). In addition, the amino acid sequences of the ORF2 of the HUN/2012/126 (GenBank accession number KX599352), GI.E/Dog/ITA/2010/Zoid (GenBank accession number JN193534) strains showed lower identities (76 and 77%, respectively) compared to the Brazilian strains (Table 2).
Phylogenetic inferences were also carried out with the partial and complete sequences of OFR2 at nucleotide and amino acid levels. The partial genome sequences of MAstV5 obtained in the present study and those available in GenBank, together with selected Mamastrovirus reference sequences from other species, generated two evolutionary trees (Fig. 2). Forty-three reference strains and the four sequences from this study corresponding to 19 Mamastrovirus species were delineated with high bootstrap support throughout the entire tree (Fig. 2A). In the MAstV5 clade, all of the present sequences clustered with “Gillingham/2012/UK” (GenBank accession number NC_026814), although with low amino acid identities of approximately 80% (Table 2). Consequently, these four new sequences grouped within Chinese sequences, suggesting a different genotype putatively named as MAsTV5a (Fig. 2B). Through a pairwise comparison of the ORF2 nucleotide sequence, a high degree of nucleotide identity, ranging from 95% to 99%, was detected among the Brazilian type strains of this study. The study sequences showed a closer relationship with the Chinese strains grouping in genotype a. More distant strains were observed among the HUN/2012/126 (GenBank accession number KX599352) Hungary strain composing the genogroup b, Bari/2008_ITA (GenBank accession number HM045005) and Gillingham/2012/UK (GenBank accession number NC_026814) strains from Italy and United Kingdom, respectively, grouping in the genotype c, and finally, forming the genotype d, HUN/2012/115 (GenBank accession number KX599351), HUN/2012/126 (GenBank accession number KX599352) strains from Hungary and GI.E/Dog/ITA/2010/Zoid (GenBank accession number JN193534) strain from Italy. This suggests a distinction of four sub-lineages among the MAstV5 species – MAstV5a to MAstV5d (Fig. 2B).
Evolutionary relationship of MAstV5 with representative MAstV genera. The percentage of replicates in which the associated virus clustered together in the bootstrap test (1000 replicates) is shown next to the branches in each tree. The trees are drawn to scale; bars represent the number of substitutions per site. All positions except ambiguous positions were included. Bootstrap values <50 were excluded. GenBank accession numbers are shown on the tree. MAstV5 sequences obtained in the present study are indicated with a black dot (●). The Kimura 2-parameter substitution model was selected for the MAstV5 ORF2 nucleotide inference, and the LG substitution model (frequencies +F) was used for the amino acid inference. The substitution-rate variation among sites was modeled with a gamma distribution (shape parameter=5). (A) Evolutionary tree based on the complete amino acid sequences of the ORF2 gene (capsid) of 47 nucleotide sequences of AstVs. (B) Evolutionary tree based on the partial nucleotide sequences of ORF2 from 30 sequences of MAstV5.
Here, in a screening of dog fecal samples, 26% (71/269) of the dogs with and without diarrhea were MAstV5 positive, as determined using RT-PCR. Likewise, non-viral agents and factors such as bacteria, intestinal parasites, malnutrition and intoxications are able to promote enteric disease mainly in the young dog population. The search for other enteric viruses in the MAstV5-positive samples from dogs with gastroenteritis showed that the dogs were also infected with other known pathogens. Moreover, we found that single MAstV5 infection was associated only with the asymptomatic state, although there is a risk that the results will be biased, since the analyzes were conducted on the basis of convenience sampling and we can not exclude the possibility that the long term of viral shedding could be an explanation for the MAstV5-positive samples detected in asymptomatic dogs, based on previous study that demonstrated the comparison between virus load and clinical manifestation26 (Fig. 1 and Table 3). These findings were not unexpected, as mixed infections are common, but more studies will be necessary to real deduce the role of MAstV5 in the cases reported here.15–24
Several reports of MAstV5 suggest a clinical association of virus molecular detection and diseased dog clinical samples.18,20–23,26,33 Furthermore, studies of the prevalence of MAstV5 in China showed that 12% (22/183) of the puppies displaying clinical signs of diarrhea were positive for MAstV5, as determined using RT-PCR, compared to none of 138 healthy dogs, although these studies did not look for other viruses that may be associated with diarrhea.19 In a study conducted in Italy, 24% of 110 stool samples collected from dogs with clinical signs tested positive for the presence of MAstV5 RNA, and 9% (10/110) of the samples showed an MAstV5-single infection, although other asymptomatic animals (9% of 75) were also positive for MAstV533. Therefore, the association with clinical signs and the shedding of the virus was described only in a case study of 2 animals, which is apparently an isolated case.26 A prevalence study in France found that 21% (66/316) of the puppies in 42% (14/33) of the breeding kennels surveyed were MAstV5 positive, as determined using RT-PCR.21 In the same report, the authors observed that puppies that were less than 7 weeks old were especially susceptible to MAstV5 infection, although a direct association with clinical signs was not possible.21 Lastly, recent studies found a MAstV5 prevalence of 6% in the United Kingdom and an infection rate of 33% in puppies under three months in Japan.22,23
The partial genomic sequencing and characterization of selected samples revealed a remarkable genetic heterogeneity of MAstV5 of Brazilian origin. Because it was hypothesized that two strains of human AstV with less than 95% identity at the nucleotide level are serologically distinguishable,34 the lower identities (<85%) shown between the capsid gene sequences analyzed here may reflect the need for a novel species classification into four genotypes – MAstV5a to MAstV5d. Additionally, phylogenetic analysis indicated that the four MAstV5 strains reported here represent a lineage that is more closely related to the Chinese strains than to the others strains, based on the high sequence identity (97%) of ORF2, according to the species demarcation criteria established by the ICTV5 (Table 2).
In summary, we found 26% MAstV5-positive fecal samples in dogs with or without gastroenteritis. Based on sequence analysis of the partial genome from four MAstV5-positive samples, we proposed a novel species classification into four genotypes – MAstV5a to MAstV5d. More studies are required to understand the biology and attempt the clinical and antigenic implications of astrovirus genotypes in dogs to elucidate the relative veterinary importance of different canine AstV types.
Conflicts of interestThe authors declare no conflicts of interest.
The authors would like to express their gratitude to the clinical practitioners who generously provided samples for analysis and to the graduate and post-graduate students of the Laboratório de Virologia for their excellent technical support in this work.