The aim of this study was to determine the association between Clostridium difficile (C. difficile) and vancomycin-resistant Enterococcus (VRE) and efficacy of screening stools submitted for C. difficile toxin assay for prevalence of VRE. Between April 2012 and February 2014, 158 stool samples submitted for C. difficile toxin to the Marmara University Microbiology Laboratory, were included in the study. Stool samples were analyzed by enzyme immuno assay test; VIDAS (bioMerieux, France) for Toxin A&B. Samples were inoculated on chromID VRE (bioMerieux, France) and incubated 24h at 37°C. Manuel tests and API20 STREP (bioMerieux, France) test were used to identify the Enterococci species. After the species identification, vancomycin and teicoplanin MIC's were performed by E test and molecular resistance genes for vanA vs vanB were detected by polymerase chain reaction (PCR). Of the 158 stool samples, 88 were toxin positive. The prevalence of VRE was 17%(n:19) in toxin positives however, 11.4% in toxin negatives(n:70). All VRE isolates were identified as Enterococcus faecium. These results were evaluated according to Fischer's exact chi-square test and p value between VRE colonization and C. difficile toxin positivity was detected 0.047 (p<0.05). PPV and NPV were 79% and 47% respectively. In our study, the presence of VRE in C. difficile toxin positives is statistically significant compared with toxin negatives (p<0.05). Screening for VRE is both additional cost and work load for the laboratories. Therefore VRE screening among C. difficile toxin positive samples, will be cost effective for determination of high risk patients in the hospitals especially for developing countries.
Colonization and infection with vancomycin-resistant Enterococcus faecium (VRE) is an increasingly common problem in hospitals of many countries worldwide and its spread is generally associated with poor hospital hygiene practice.
Risk factors for VRE colonization include host characteristics (immunosuppression, neutropenia, and renal insufficiency), hospital factors (admission to an intensive care unit (ICU) or oncology ward, proximity to a VRE-colonized patient, and extended length of stay), and antimicrobial use.1–3
VRE colonization increases the patient risk of developing infections, such as bloodstream infections. Rapid and accurate identification of VRE is crucial in the management and treatment of both colonized and infected patients and to prevent the nosocomial spread of this resistant bacteria. Therefore screening for VRE from rectal swabs is a routine procedure in most hospitals. However, routine screening for VRE represents a financial burden for hospitals, mainly in the developing countries. Therefore, it is important to select appropriate patients for this screening especially in the high risk wards.4
Clostridium difficile is anaerobic bacteria that produce multiple toxins including A and B toxins (A&B toxins) and cause diarrhea and inflammation in infected patients. VRE and C. difficile are both nosocomial pathogens and thus have similar risk factors including antibiotic treatment and hospitalization.5
In this study we aimed to assess the prevalence of gastrointestinal colonization by VRE in stool samples submitted for C. difficile toxin testing and to assess the cost and advantages of this laboratory-based surveillance.
Materials and methodsSamplesStools submitted for C. difficile toxin testing between April 2012 and February 2014, at the Marmara University Microbiology Laboratory, were included in the study.
C. difficile toxin detectionStool samples were analyzed using an enzyme-linked fluorescent (ELFA) assay, VIDAS®C. difficile Toxin A & B (BioMérieux, France). Stools were mixed with 200μL distilled water and centrifuged at 12,000×g. Supernatant (300μL) was loaded in to the test well. After 75min, test results were evaluated as <0.13-negative, ≥0.13-<0.37-intermedate, ≥0.37 positive.
Enterococcus spp. identificationSamples were inoculated on chromID VRE (BioMérieux, France) and incubated at 37°C in normal atmosphere and examined for growth after 24–48h. Purple colonies on chromID VRE were presumptively identified as VRE. After Gram staining, positive cocci were then subcultured to sheep blood agar and incubated at 37°C in normal atmosphere, and examined after 24h. In addition to colony morphology and Gram staining, catalase reaction, growth in 6.5% NaCl and pyrrolidonyl aminopeptidase activity were used for the identification at genus level.6 Additionally, API20 STREP (BioMérieux, France) test was used to identify the enterococci at species levels.
Vancomycin resistance detectionAfter species identification, vancomycin and teicoplanin minimal inhibitory concentrations (MICs) were determined for enterococci using Etest method (Biomérieux, France). MICs were interpreted using the following breakpoints according to CLSI standards: Vancomycin ≤4μg/mL (susceptible), 8–16μg/mL (intermediate), ≥32μg/mL (resistant); and teicoplanin ≤8μg/mL (susceptible), 8–16μg/mL (intermediate), ≥32μg/mL (resistant).7
Detection of the genes vanA and vanBThese genes were detected by multiplex PCR-based test. Two or three colonies of VRE were suspended in distilled water and centrifuged at 10,000×g. The suspension was then boiled at 100°C for 10min. Then 2.5μL supernatant was added to PCR Master Mix (Promega), primers(vanA, vanB) to make a final volume of 25μL. Primers for van; vanA1, vanA2 primers (5′ GGGAAAACGACAATTGC 3′ and5′ GTACAATGCGGCCGTTA 3′, 732bp) and for vanB vanB1, vanB2 primers (5′ ATGGGAAGCCGATAGTC 3′ and 5′ GATTTCGTTCCTCGACC 3′, 635bp) were used.8 PCR amplification was performed by initial denaturation 94°C for 2min, denaturation at 94°C for 1min, annealing at 54°C for 1min, extension at 72°C for 1min (30 cycles). PCR products were loaded in 1.5% agarose gel and stained by ethidium bromide and visualized under ultraviolet transiluminator.
Statistical methodsAll results were evaluated using SPSS 17.0 version and analyzed according to Fischer's Exact Chi-Square test and p<0.05 was accepted as significant.
ResultsBetween August 2012 and February 2014, 158 stool specimens submitted for C. difficile toxin A assay were tested for VRE in the Microbiology Laboratory of Marmara University Hospital. C. difficile infection is the most important cause of nosocomial diarrhea, and the use of antibiotics have been implicated as a major risk factor for this condition. In our study, 113 (71.5%) of the 158 patients were hospitalized and treated with antibiotics. C. difficile toxin was detected in 88 (55.7%) specimens.
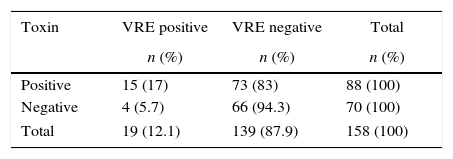
Of the 88 toxin positive samples, 27 (30.6%) were positive for enterococci whereas of the 70 negative samples, 8 (11.4%) were positive for these bacteria. A total of 35 Enterococcus spp. were isolated in the specimens. thirty-two of these isolates were identified as E. faecium (91.4%) and three as E. faecalis (8.6%).
Of the 35 isolates 19 (54.2%) were vancomycin and teicoplanin resistant. All the resistant isolates were E. faecium and they carried van A gene. van B gene was not detected (Fig. 1).
The vancomycin MICs for the enterococcal isolates were ≥256 whereas teicoplanin MICs were 32–256μg/mL. Etest MIC results showed 100% agreement with vanA PCR data. VRE was isolated from 15 (17%) of 88 C. difficile toxin positive specimens, compared with 4 (5.7%) of 70 C. difficile toxin negative specimens (Table 1).
Positive predictive value (PPV) and negative predictive value (NPV) were 79% and 47% respectively. These results were evaluated according to Fischer's exact chi-square and p value between VRE colonization and C. difficile toxin positivity was found to be 0.047 (p<0.05). Patients whose stool specimens were positive for C. difficile toxin A were significantly more likely than those whose specimens were negative to have VRE detected.
DiscussionFrom the early 1970s Enterococcus spp. became one of the most common pathogens in nosocomial infections. These bacteria are commonly resistant to most antibiotics.1–3 Because the increased VRE incidence, rapid identification methods are getting importance, specially for inpatients. Chromogenic medium is one of these rapid methods both for identification and species isolation. ChromID VRE (bioMerieux, France) medium containing 8μg/mL vancomycin is a selective for the identification of VRE. Sensitivity and specificity of the medium is high.9,10 In the study of Ledeboer et al., 120 stool samples were inoculated on chromID VRE and the sensitivity of identification rates of E. faecium and E. faecalis were 85.4% and 90.0% respectively whereas specificity was 100% in both species.11 Malignity, chronic kidney failure, neutropenia, transplantation chemotherapy are risk factors for VRE infection. Similar risk factors are also important for C. difficile infections. Gerding et al. first detected the relationship between VRE detection and C. difficile toxin positives.12 Garbutt et al. studied the presence of C. difficile toxin in 215 patient samples and detected VRE in 41(19%) of these.13 In another study, Shay et al. detected 10 (21.7%) C. difficile infection in the 46 VRE patient with blood stream infection (BSI).14 Jordens et al. found this ratio as 11% whereas Rafferty et al. detected 16.5%.15,16 Leber et al. evaluated 50 stool samples and detected VRE colonization in 3.2% of the specimens submitted for C. difficile toxin assay.17
In our study, VRE was detected in 17% of the C. difficile toxin positive samples. Our results were very similar to those obtained by Garbutt et al. and Rafferty et al.13,16 Similarly, Fujitani et al. detected VRE colonization in 88 of the 158 C. difficile toxin positive patients.18 VRE colonization was detected as 5.7% in toxin negatives. Presence of VRE in toxin positives are statistically significant comparing to toxin negatives (p<0.05). All VRE isolates had vanA gene, none vanB gene was detected. vanB gene is a rare resistance gene for our country.
Vidas C. difficile toxin assay is a rapid and relatively simple test; however it has low sensitivity. The sensitivity, specificity and positive and NPV using RT-PCR kit were 100%, 98.3%, 84.6% and 100%, respectively, while using Vidas C. difficile toxin assay were 63.6%, 100%, 100% and 96.6%, respectively.19 However, RT-PCR is expensive and needs experienced staff.
During our study period, VRE isolation ratio were 12.9% (n:486) in 3772 rectal swabs in our laboratory. C. difficile toxin positivity was 3.8%, however VRE positivity in C. difficile toxin positive samples were 17%. Therefore, these patients should be considered at great risk to be colonized by VRE. Gastrointestinal system is one of the most important source of VRE. To detect and isolate colonizated patients, is recommended to prevent transmission of VRE. However, routine screening for VRE causes staff workload and increases cost for both hospitals and laboratories. VRE screening among C. difficile toxin positive samples can be cost effective and efficient strategy especially for developing countries. In a study, from our country, the estimated costs for screening VRE periodically were $19,074 annually.20 Screening VRE from stool samples submitted for C. difficile toxin assay eliminates the need for a separate specimen collection, which may be a source of stool contaminations.21 It is also less invasive procedure comparing to rectal swabs. In developing countries, like our country, hospitals should determine their high risk clinics for VRE to limit their screening priority. Appropriate choice of high risk patients like C. difficile toxin positives, will lead the screening to be more cost effective. We finally suggest that among the C. difficile toxin negative patients, those at increased risk for VRE colonization, such as previously being colonized or infected with VRE, being transferred from hospital with VRE outbreak or high VRE colonization or infection rates on admission, should also be tested for VRE.
Conflicts of interestThe authors declare no conflicts of interest.
This study was supported by a grant from Marmara University Scientific Research Commission, with a grant number of SAG-C-YLP-130213-0025.