Carbapenems are considered last-line agents for the treatment of serious infections caused by Klebsiella pneumoniae, and this microorganism may exhibit resistance to β-lactam antibiotics due to different mechanisms of resistance. We evaluated 27 isolates of K. pneumoniae resistant to carbapenems recovered from inpatients at the University Hospital of Santa Maria-RS from July 2013 to August 2014. We carried out antimicrobial susceptibility, carbapenemase detection, testing for the presence of efflux pump by broth microdilution and loss of porin by sodium dodecyl sulfate polyacrylamide gel electrophoresis. Genetic similarity was evaluated by ERIC-PCR. High levels of resistance were verified by the minimum inhibitory concentration for the antimicrobials tested. The blaKPC gene was present in 89% of the clinical isolates. Blue-Carba and combined disk with AFB tests showed 100% concordance, while the combined disk test with EDTA showed a high number of false-positives (48%) compared with the gold-standard genotypic test. Four isolates showed a phenotypic resistance profile consistent with the overexpression of the efflux pump, and all clinical isolates had lost one or both porins. The ERIC-PCR dendrogram demonstrated the presence of nine clusters. The main mechanism of resistance to carbapenems found in the assessed isolates was the presence of the blaKPC gene.
Cephalosporins used to be the antibiotics commonly prescribed for treatment of severe Klebsiella pneumoniae infections. However, due to the high frequency of extended spectrum β-bactamases (ESBL) producing K. pneumoniae, carbapenems have become the most common antibiotics prescribed for the treatment of these infections. Unfortunately, the high consumption of carbapenems has been accompanied by the emergence and spread of carbapenem-resistant K. pneumoniae.1
Resistance to carbapenems can be associated with the production of carbapenemases, loss of porins and overexpression of the efflux pump. K. pneumoniae have acquired genes encoding for carbapenemases, which are enzymes capable of breaking down most β-lactams antibiotics, including carbapenems, and thus conferring resistance to these drugs, which can result in treatment failure.2 The carbapenemases are classified in A, B and D Ambler classes. The most common carbapenemase class A is K. pneumoniae carbapenemase (KPC), the most common carbapenemase class B is New Delhi metalo β-lactamase (NDM) and the most common carbapenemase class D is Oxacillinase-48 (OXA-48), occurring in K. pneumoniae.3
K. pneumoniae presents two main porins, OmpK35 and OmpK36. The loss of these porins (OmpK35 and OmpK36) may play an important role in the development of resistance to carbapenems in K. pneumoniae.4 Furthermore, overexpression of the AcrAB efflux pump has also been proposed as being responsible for reduced susceptibility to ertapenem and meropenem antibiotics in some strains.5
The aim of this study was to evaluate mechanisms involved in carbapenem resistance among K. pneumoniae isolates. As carbapenemases are most often acquired by horizontal transfer between strains, whereas change in carbapenem permeability or efflux is a result of point mutations, it is important to study these different resistance mechanisms in locally collected strains, as these resistance mechanism profiles might vary temporally and geographically.
Materials and methodsBacterial strainsTwenty-seven non-duplicated K. pneumoniae clinical isolates resistant to carbapenems collected from July 2013 to August 2014 at the Santa Maria University Hospital, Brazil were included in this study. Species identification was carried out on the Vitek® 2 automated identification system (Biomérieux, France) and confirmed with MALDI-TOF MS in a Microflex LT apparatus (Bruker Daltonics, Germany) considering a score value between 2.0 and 2.299.6
Susceptibility testingThe antimicrobials used for susceptibility testing were imipenem, meropenem, ertapenem, cefepime, ceftazidime, and cefoxitin. The tests were performed by broth microdilution according to CLSI guidelines.7Escherichia coli ATCC 25922 was used as a quality control.
CarbapenemasesThe detection of genes blaKPC,8blaOXA-489 and blaNDM10 was carried out using the multiplex PCR technique, as well as the detection of genes blaSIM, blaSPM, blaIMP, blaVIM and blaGIM.11 The detection of blaGES was carried out with simplex PCR.12
Disk diffusion assay using phenyl boronic acid, EDTA and cloxacillinFor phenotypic detection of the KPC- and metallo-β-lactamases (MBLs), we used disk diffusion assays with phenylboronic acid (AFB)13 and ethylenediamine tetra-acetic acid (EDTA), respectively.14 In both tests, an increase of 5mm in zone diameter in the presence of AFB or EDTA compared with either meropenem or imipenem tested alone was considered to represent a positive result for the presence of KPC β-lactamase or MBL enzyme, respectively.
For the detection of plasmid-mediated AmpC, we used test results from AFB and cloxacillin (CLOXA)13 in combination with meropenem and imipenem, compared with carbapenem disks alone. An increase of 5mm in zone diameter for both AFB and CLOXA was considered to be a positive test result for the presence of AmpC. Strains KPC2-K. pneumoniae, IMP1-K. pneumoniae and E. coli ATCC 25922 were used as quality controls.
Blue-Carba testThe Blue-Carba test consists of the detection of hydrolysis of the carbapenem β-lactam ring in a bacterial extract through the acidification of bromothymol blue indicator. A loop of a pure bacterial culture was directly suspended in 100μL of the test solution (aqueous solution of bromothymol blue, ZnSO4 and imipenem) and in the negative-control solutions (without imipenem), followed by incubation at 37°C for 2h, with the first reading at 15min. Carbapenemase activity was revealed when the test and negative-control solutions were (i) yellow versus blue, (ii) yellow versus green, or (iii) green versus blue. Noncarbapenemase producers remained blue or green on both solutions. Strains harboring the genes blaKPC, blaIMP and blaOXA-48 were used as positive controls, and E. coli ATCC 25922 was used as a negative control.15
Efflux pumpThe technique was carried out by broth microdilution using antimicrobial alone and associated with the efflux pump inhibitor carbonyl cyanide m-cholorophenyl hydrazone (CCCP), using half of the minimal inhibition concentration (MIC) for CCCP. The reduction in MIC of the antimicrobial sample, associated with the efflux pump inhibitor, was indicative of efflux pump overproduction.16
OMP analysisOuter membrane protein (OMP) profiles were analyzed by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE).17 OmpK35 of K. pneumoniae, OmpK36 of K. pneumoniae and OmpK35 and OmpK36 of K. pneumoniae were used as control strains belonging to the Alerta Laboratory (UNIFESP).
Enterobacterial repetitive intergenic consensus sequences (ERIC-PCR)ERIC-PCR was performed using 12.5μL of MasterMix SYBRGreen (1×), 1μL of DMSO (dimethyl sulfoxide), 2.5μL of template, 1μL from primer ERIC2 (AAGTAAGTGACTGGGGTGAGCG)18 and water to complete the total volume of 25μL. The PCR amplifications were performed in a thermocycler as follows: 94°C for 10min and 40 cycles of 30s at 92°C, 1min at 52°C and 8min at 65°C. A final extension step of 16min at 65°C was performed. ERIC-PCR fingerprint analysis was performed with BioNumerics 6.6 software (Applied Maths, Belgium), based on Dice's similarity coefficient and using unweighted-pair group method with arithmetic mean (UPGMA). Band position tolerance was 2.0%, and optimization was 0.8%. Isolates with a similarity coefficient ≥90% were considered to belong to the same cluster.19
ResultsTwenty-four (89%) isolates were positive for the blaKPC gene. The other carbapenemase genes investigated were not detected in the isolates. Among the KPC producing K. pneumoniae, the values of MIC50 and MIC90 demonstrate a high level of resistance for all antimicrobials tested (Table 1).
MIC 50 and 90 for antimicrobials tested against 27 clinical isolates of Klebsiella pneumoniae.
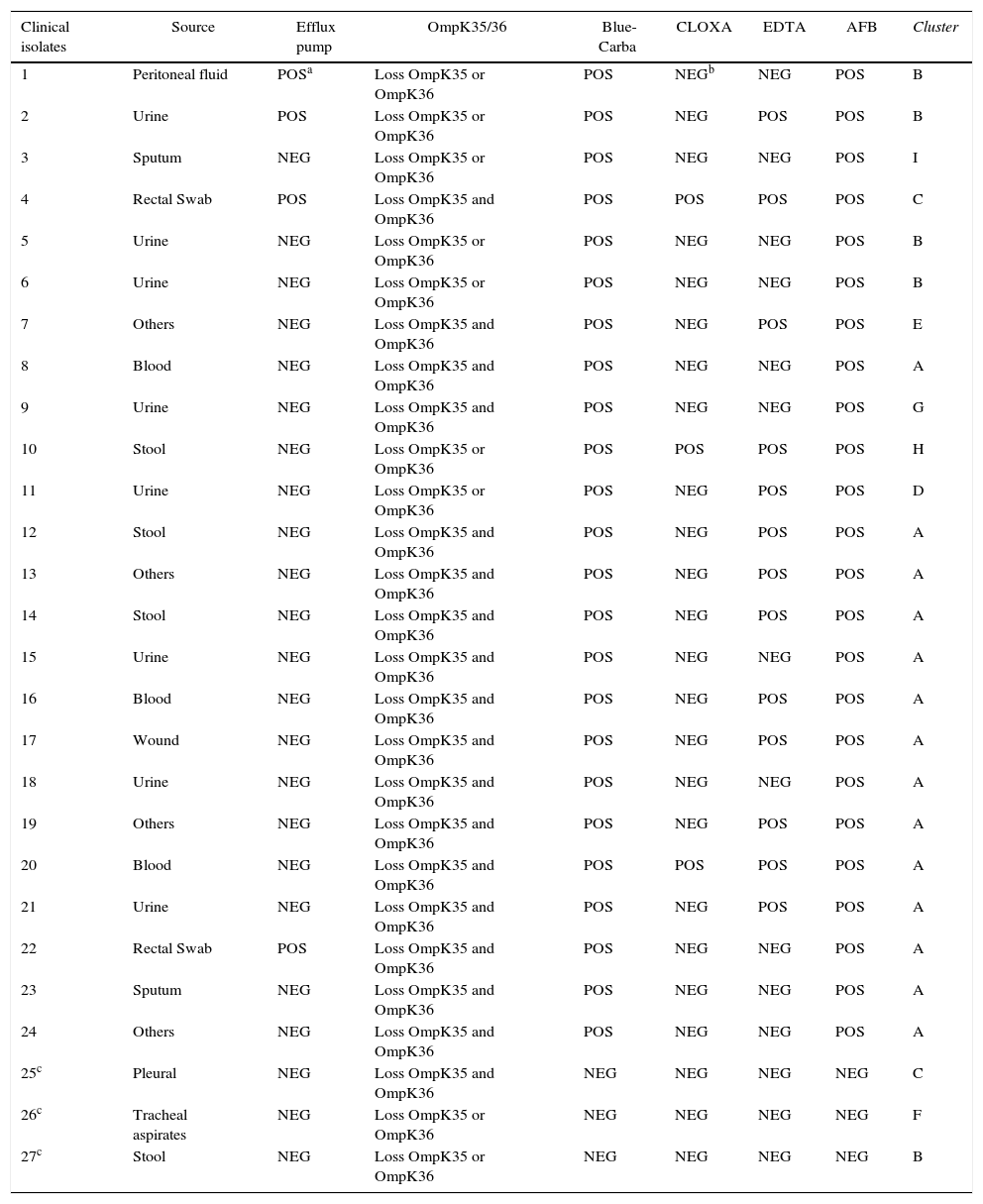
Tests for carbapenemase phenotypical and molecular detection, as well as overproduction of the efflux pump and porin loss, are shown in Table 2.
Phenotypic and genotypic characterization of the 27 Klebsiella pneumoniae isolates.
| Clinical isolates | Source | Efflux pump | OmpK35/36 | Blue-Carba | CLOXA | EDTA | AFB | Cluster |
|---|---|---|---|---|---|---|---|---|
| 1 | Peritoneal fluid | POSa | Loss OmpK35 or OmpK36 | POS | NEGb | NEG | POS | B |
| 2 | Urine | POS | Loss OmpK35 or OmpK36 | POS | NEG | POS | POS | B |
| 3 | Sputum | NEG | Loss OmpK35 or OmpK36 | POS | NEG | NEG | POS | I |
| 4 | Rectal Swab | POS | Loss OmpK35 and OmpK36 | POS | POS | POS | POS | C |
| 5 | Urine | NEG | Loss OmpK35 or OmpK36 | POS | NEG | NEG | POS | B |
| 6 | Urine | NEG | Loss OmpK35 or OmpK36 | POS | NEG | NEG | POS | B |
| 7 | Others | NEG | Loss OmpK35 and OmpK36 | POS | NEG | POS | POS | E |
| 8 | Blood | NEG | Loss OmpK35 and OmpK36 | POS | NEG | NEG | POS | A |
| 9 | Urine | NEG | Loss OmpK35 and OmpK36 | POS | NEG | NEG | POS | G |
| 10 | Stool | NEG | Loss OmpK35 or OmpK36 | POS | POS | POS | POS | H |
| 11 | Urine | NEG | Loss OmpK35 or OmpK36 | POS | NEG | POS | POS | D |
| 12 | Stool | NEG | Loss OmpK35 and OmpK36 | POS | NEG | POS | POS | A |
| 13 | Others | NEG | Loss OmpK35 and OmpK36 | POS | NEG | POS | POS | A |
| 14 | Stool | NEG | Loss OmpK35 and OmpK36 | POS | NEG | POS | POS | A |
| 15 | Urine | NEG | Loss OmpK35 and OmpK36 | POS | NEG | NEG | POS | A |
| 16 | Blood | NEG | Loss OmpK35 and OmpK36 | POS | NEG | POS | POS | A |
| 17 | Wound | NEG | Loss OmpK35 and OmpK36 | POS | NEG | POS | POS | A |
| 18 | Urine | NEG | Loss OmpK35 and OmpK36 | POS | NEG | NEG | POS | A |
| 19 | Others | NEG | Loss OmpK35 and OmpK36 | POS | NEG | POS | POS | A |
| 20 | Blood | NEG | Loss OmpK35 and OmpK36 | POS | POS | POS | POS | A |
| 21 | Urine | NEG | Loss OmpK35 and OmpK36 | POS | NEG | POS | POS | A |
| 22 | Rectal Swab | POS | Loss OmpK35 and OmpK36 | POS | NEG | NEG | POS | A |
| 23 | Sputum | NEG | Loss OmpK35 and OmpK36 | POS | NEG | NEG | POS | A |
| 24 | Others | NEG | Loss OmpK35 and OmpK36 | POS | NEG | NEG | POS | A |
| 25c | Pleural | NEG | Loss OmpK35 and OmpK36 | NEG | NEG | NEG | NEG | C |
| 26c | Tracheal aspirates | NEG | Loss OmpK35 or OmpK36 | NEG | NEG | NEG | NEG | F |
| 27c | Stool | NEG | Loss OmpK35 or OmpK36 | NEG | NEG | NEG | NEG | B |
The 27 isolates of K. pneumoniae were grouped in nine different clusters, designated A, B, C, D, E, F, G, H and I as determined by ERIC-PCR (Fig. 1). The major group is A that included 14 isolates (52%), followed by group B with five isolates (19%) and group C with two isolates (7.4%). Clusters D, E, F, G, H and I had one isolate each.
DiscussionThe class A K. pneumoniae carbapenemase (KPC) is one of the most common mechanisms of carbapenem resistance in Enterobacteriaceae. The KPC enzymes confer resistance to all β-lactam agents, including penicillins, cephalosporins, monobactams, and carbapenems.20 In our study, when comparing carbapenem resistance level between the group that harbored the blaKPC gene (group KPC) and isolates that tested negative for its presence (group non KPC), it was determined that the KPC enzyme probably could represent the main mechanism responsible for the high level of resistance to carbapenems observed among the isolates. All isolates presented a loss of one or both porins; therefore, this resistance mechanism can contribute to the high level of resistance observed in the non KPC group. The presence of efflux pump in some isolates of the KPC group did not change the high level of antibiotic resistance (data not shown).
The analysis of ERIC-PCR band patterns revealed a clonal spread at our institution. This finding is actually a global concern and is well documented in many hospitals, including other Brazilian hospitals.21–23 It will be interesting to submit the isolates to further analysis by pulsed-field electrophoresis and multilocus sequence typing to confirm the clonal relatedness observed among the carbapenem-resistant K. pneumoniae.
Methods for the phenotypic identification of carbapenemases are based on the use of specific inhibitors (AFB and EDTA) or in the detection of hydrolysis of the carbapenem β-lactam ring with an indicator. Both the phenotypic AFB-based method and Blue-Carba were 100% sensible and specific, considering PCR as the gold standard. Studies have shown that the sensitivity of the disk diffusion assay using AFB is 100% 14,24 and the Blue-Carba test presents 100% specificity and sensitivity.15
The MBLs phenotypic detection test using EDTA presented a high false-positive rate (48%) when compared with the PCR technique, a fact that has also been reported in the literature.15,25–27 These results can be explained based on the effect on membrane permeability, which can increase the susceptibility to many antimicrobial agents, including imipenem and meropenem.26,27 It can also be associated with a low production of β-lactamase or even with production of some unknown enzyme.28 The high false-positive rates observed in this study could be due to the clonal nature of our isolates, as 61.5% of them belonged to cluster A. However, we found false-positive results among isolates belonging to clusters B, C, D, E and H.
In our collection, the absence of one or both porins (OmpK35 and OmpK36) was found in all the isolates. Due to the limitation of the SDS-PAGE technique and the very close molecular masses of the porins evaluated, we were not able to distinguish exactly which of the porins were absent.
The loss of the major porins OmpK35/36 is often observed in K. pneumoniae clinical isolates resistant to carbapenems, demonstrating an increase in MICs to cephalosporins and carbapenems. The loss of only the OmpK35 porin did not significantly influence the resistance to carbapenems.29,30 In contrast, the loss of OmpK35/36 conferred 31-, 8- and 4-fold increases in the MIC of ertapenem, meropenem and doripenem, respectively, and led to ertapenem resistance.1 This suggests that when a clinical isolate from our study lost a porin, it was most likely OmpK36. However, additional molecular analysis will be needed to confirm this result. Clinical isolates tested against cephalosporins and carbapenems showed resistance, and the highest MIC values (>128μg/mL) were associated with the loss of both porins (data not shown).
In our study, the active efflux mechanism was always associated with other mechanisms that had already been proven to participate in the resistance to carbapenems. This active efflux was found in isolates from clusters A, B and C, and thus further studies should be carried out to evaluate the overexpression and characterization of efflux pumps in these clusters. Nevertheless, the literature is controversial regarding the participation of active efflux in the resistance to carbapenems.31
In conclusion, the major mechanism related to carbapenem resistance that we found was the presence of the blaKPC gene associated with porin loss. Different resistance mechanisms to carbapenems were present in clinical isolates, demonstrating that often the cause of resistance cannot be only a single mechanism but the combination of many. Our results showed that 14% of clinical isolates exhibit a combination of three different resistance mechanisms (carbapenemases, drug efflux and loss of porin). The detection of these mechanisms becomes extremely important for the implementation of infection control and prevention measures as well as epidemiological surveillance.
Conflicts of interestAna Cristina Gales has recently received research funding and/or consultation fees from AstraZeneca, MSD and Novartis. All the other authors declare no conflicts of interest.
Coordenação de Aperfeiçoamento de Pessoal do Ensino Superior (CAPES) by the Masters degree grant and Dr. Anna Sara Levin (Laboratório Investigação Médica-LIM-54 – Faculdade de Medicina da Universidade Federal de São Paulo) for supplying positive control strains.