Salmonella is recognized as a common foodborne pathogen, causing major health problems in Saudi Arabia. Herein, we report epidemiology, antimicrobial susceptibility and the genetic basis of resistance among S. enterica strains isolated in Saudi Arabia. Isolation of Salmonella spp. from clinical and environmental samples resulted in isolation of 33 strains identified as S. enterica based on their biochemical characteristics and 16S-rDNA sequences. S. enterica serovar Enteritidis showed highest prevalence (39.4%), followed by S. Paratyphi (21.2%), S. Typhimurium (15.2%), S. Typhi and S. Arizona (12.1%), respectively. Most isolates were resistant to 1st and 2nd generation cephalosporin; and aminoglycosides. Moreover, several S. enterica isolates exhibited resistance to the first-line antibiotics used for Salmonellosis treatment including ampicillin, trimethoprim–sulfamethoxazole and chloramphenicol. In addition, the results revealed the emergence of two S. enterica isolates showing resistance to third-generation cephalosporin. Analysis of resistance determinants in S. enterica strains (n=33) revealed that the resistance to β-lactam antibiotics, trimethoprim–sulfamethoxazole, chloramphenicol, and tetracycline, was attributed to the presence of carb-like, dfrA1, floR, tetA gene, respectively. On the other hand, fluoroquinolone resistance was related to the presence of mutations in gyrA and parC genes. These findings improve the information about foodborne Salmonella in Saudi Arabia, alarming the emergence of multi-drug resistant S. enterica strains, and provide useful data about the resistance mechanisms.
Although the high advances in safety measures taken in food and drinking water, Salmonella infections (Salmonellosis) are still recognized as one of most global foodborne diseases with a wide range of hosts. Salmonella spp. is facultative anaerobic intracellular gram negative flagellated bacilli that belong to family Enterobacteriaceae; and the genus consists of two main species; S. bongori and S. enterica.1 The causative agent of salmonellosis is S. enterica subsp. enterica, which is subdivided into more than 2500 serovars based on antigenic differences in the lipopolysaccharide O antigen and two flagellin structures, most of them are recognized as potential human pathogen.2Salmonella infections are divided into two main types including (i) invasive typhoidal salmonellosis that caused by S. enterica serotype Typhi and Paratyphi A, B and C causing enteric fever gastroenteritis and bacteremia; (ii) non-typhoidal salmonellosis (NTS) caused by S. enterica serotype Enteriditis and S. enterica serotype Typhimurium, which have a broad vertebrate hosts range and cause various symptoms that usually include diarrhoeal disease.3,4 Although, typhoidal Salmonella caused severe and life-threatening diseases, the non-typhoidal Salmonella is associated with self-limiting diseases such as gastroenteritis, with more severe cases reported in immunocompromised individuals.5 Generally, Salmonella infections are transmitted to human via consumption of contaminated water and food particularly the animal products, however typhoidal Salmonella, which is restricted to human, is transmitted by fecal oral route or direct contact with the infected persons.6
Recently, the selective pressure owing to the misuse of antimicrobial agents in humans and domestic animals led to the emergence of multidrug-resistant S. enterica strains, including resistance to quinolone, fluoroquinolones and the third generation of cephalosporin which are the current drugs of choice for salmonellosis treatment in severe cases, representing a significant public health problem throughout the world.7 There are several evidences underpin that the antibiotics resistance among Salmonella strains is attributed to intensive use of antibiotics as growth promoters in animals feeding.8 Moreover, the intensive use of antimicrobial agents to treat both animal and human infections led to flourish the horizontal resistance genes transfer between bacterial communities.9 The antimicrobial resistance in S. enterica is attributable to various mechanisms such as enzymatic degradation of some antimicrobial agents, blocking the cell permeability to antibiotics, activation of antimicrobial efflux pumps, and alteration the site of drugs actions.7 The aims of this study were to determine the predominant serotype of Salmonella isolated in Saudi Arabia, emergence of antibiotics resistance among Salmonella strains and to investigate the genetic basis of antimicrobial resistance among the isolates.
Materials and methodsClinical samples collectionDifferent clinical specimens were collected from patients with symptoms suspected to be Salmonella infection (King Khalid University Hospital, Riyadh, Saudi Arabia). The clinical samples included stools, urine and blood samples. In addition, various samples were collected from Sewage Treatment Plant in Riyadh (Saudi Arabia). The specimens were collected under sterile conditions and transferred to the laboratory in cold box within 1–2h for bacterial isolation.
Bacterial isolation and identificationSerial dilutions (10-fold) of the clinical and environmental samples were made in 1% sterile peptone water (Difco, UK). Then 0.1mL of each dilution was inoculated into Salmonella selective medium, Selenite F broth (Oxoid, UK), to enhance the growth of Salmonella spp. and inhibit the other contaminants, and incubated at 37°C for 24–48h. After enrichment, the growth was transferred to the media recommended for Salmonella spp. including: Xylose lysine deoxycholate agar (XLD) (Oxoid, UK) and Deoxycholate citrate agar (DCA) (Oxoid, UK), and incubated at 37°C for 24h.9Salmonella colonies, characterized by producing non-lactose fermenting pale colored colonies with black centers on DCA medium and pink-red colonies with black centers on XLD medium, were picked up and sub-cultured several times on fresh plates until homogeneous colonies were obtained. The colonies were confirmed as Gram negative bacteria using Gram staining procedures, and glycerol cultures of all of the isolates were prepared and stored at −80°C for further analysis. The isolated bacterial strains were subjected to identification using biochemical tests and Vitek® 2-C15 automated system for bacterial identification (BioMerieux Inc., France), according to manufacturer's instructions. Furthermore, bacterial identification was confirmed by 16S rDNA sequencing analysis.
16Sr rDNA sequencing analysisThe Salmonella isolates (n=33) were inoculated into nutrient broth (Merck, UK) and incubated at 37°C for 18h. Total bacterial DNA was extracted using DNeasy Blood & Tissue Kits (Qiagen, UK) according to the manufacturer's instructions. The 16S rDNA genes of the isolated Salmonella spp. strains (n=33) were PCR-amplified using the universal eubacterial primers10: 16F27 (5′-AGA GTT TGA TCC TGG CTC AG-3′) and 16R1525 (5′-AAG GAG GTG ATC CAG CCG CA-3′). The PCR amplification was performed using purified genomic DNA of the Salmonella spp. strains (n=33) as templates. The PCR reaction (50μL) contained PCR master mix (Promega, USA) (14μL), forward primer (4μL), reverse primer (4μL), DNA templates (4μL), and nuclease-free water (13μL). The PCR reaction was carried out under the following conditions: initial denaturation for 5min at 95°C, followed by 35 cycles of denaturation at 95°C (30s); annealing at 52°C (30s); extension at 70°C (1.5min), and then, a final extension step at 70°C (5min). The PCR products were analyzed by 1% (w/v) agarose gel electrophoresis using a 1kbp DNA ladder (Qiagen, UK) as molecular size standard. The amplified 16S rDNA products were purified from the agarose using QIAquick gel Extraction Kits (Qiagen, UK). The purified 16S rDNA amplicons were sequenced by an automated sequencer (Macrogen, Korea) using the 16F27 and 16R1525 primers mentioned above. BLAST analysis of the obtained sequences was performed by NBCI online database to determine the phylogenetic grouping of the isolated strains (http://www.ncbi.nlm.nih.gov/genbank/index.html).
Salmonella serotypingThe serotyping of the isolated Salmonella (n=33) strains were carried out according to Kauffman–White Scheme11 by slide agglutination tests using commercially available mono- and poly-O groups Salmonella A, B, C, D, E antisera (Remel, Europe Ltd., UK). In addition, polyvalent Salmonella antisera phase 1 and phase 2 flagellar H antigens were used for serovars determination of the isolated Salmonella. Briefly, a loopful of each isolate grown on Brain Heart Infusion (BHI) agar was suspended in 50μL of sterile distilled water on a glass slide, and then mixed with one drop of each antiserum. The slide was rotated gently for 1min, and observed for appearance of any agglutination reaction using indirect lighting over a dark background. However, some strains (S. Typhi and S. Paratyphi C) may possess capsular polysaccharide antigen, known as Vi, that render the strains non-agglutinable in O-antisera. Therefore, the O-antigen was detected after destruction of Vi antigen by boiling the culture for 10min. E. coli cell suspension was used as negative control.
Antimicrobial susceptibility testingThe isolates identified as Salmonella (n=33) were tested for their susceptibility to 26 commonly used antimicrobial agents using disk diffusion assay and Vitek® 2-C15 automated system. The tested antibiotics included (Oxoid Limited Company, UK): kanamycin (k), tetracycline (TE), streptomycin (S), erythromycin (E), neomycin (N), ampicillin sulbactam (SAM), chloramphenicol (C), amikacin (AN), amoxicillin/clavulanic acid (AMC), ampicillin (AM), cefalotin (CF), cefepime (FEP), cefotaxime (CTX), cefoxitin (FOX), cefpodoxime (CPD), ceftazidime (CAZ), cefuroxime (CXM), ciprofloxacin (CIP), gentamicin (GM), meropenem (MEM), nitrofurantoin (FT), norfloxacin (NOR), piperacillin (PIP), piperacillin/tazobactam (TZP), tobramycin (TM) and trimethoprim/sulfamethoxazole (SXT). For disk diffusion assay, the bacterial strains were sub-cultured on fresh Mueller–Hinton agar plates (Difco, UK) for 24h at 37°C. After the incubation period, the cells were harvested using a sterile loop and suspended in sterile saline solution to be equivalent to 0.5 McFarland standards. The cell suspensions were inoculated onto Mueller–Hinton agar plates using sterile cotton swabs, and various antibiotic discs were placed on the agar plate surfaces and incubated for 24–48h at 37°C.12 The results were interpreted Clinical and Laboratory Standard Institute (CLSI) guidelines.13Escherichia coli ATCC 25922, E. coli ATCC 35218 and Pseudomonas aeruginosa ATCC 27853 were used as control organisms.
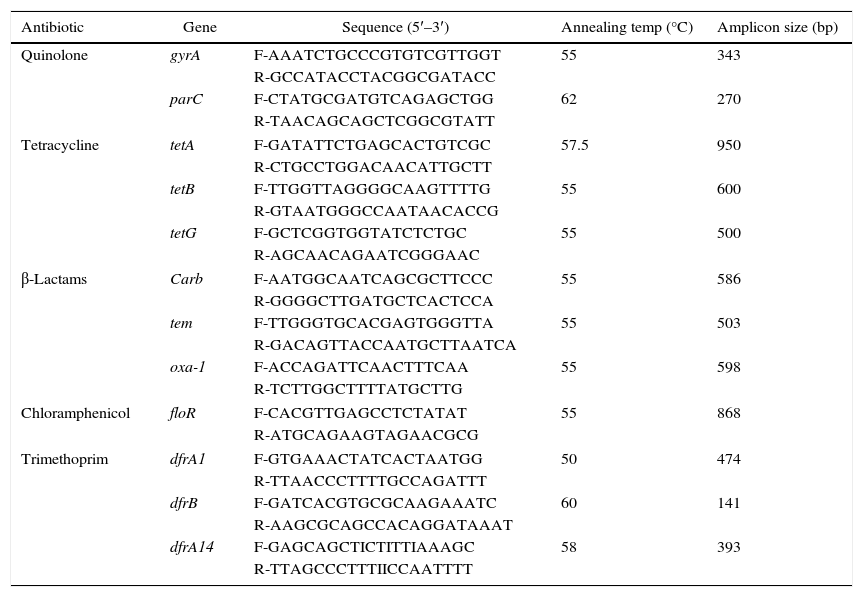
Detection of antimicrobial resistance determinantsPolymerase chain reaction (PCR) was used for detection of various antibiotics resistance genes (n=12) in the isolated S. enterica strains (n=33) according to previously reported method with some modifications.14 The isolates were tested for the presence of the carb-like, tem, and oxa-1 genes, encoding resistance to beta-lactams antibiotics; floR gene for chloramphenicol resistance; tetA, tetG, and tetB encoding resistance to tetracycline; dfrB, dfrA1 and dfrA14 genes encoding trimethoprim resistance; and mutation in gyrA and ParC for fluoroquinolone resistance. PCR amplification of the resistance genes was carried out using a list of specific primers shown in Table 1.14 DNA amplification was carried out in PCR thermocycler (Biotech prime thermocycler UK), with the following reaction conditions: initial denaturation for 2min at 94°C, followed by 35 cycles of denaturation for 1min at 94°C, 30s at annealing temperature of each primer, and extension at 72°C for 1.5min and a final extension for 5min at 72°C. The amplified genes were analyzed by 1.5–2% agarose gel electrophoresis. In addition, the PCR products of gyrA and parC genes were purified from gels by using QIAquick gel extraction kit (Qiagen, UK); the genes were sequenced by automated sequencer services (Macrogen, Korea), and aligned with the known genes available in NBCI online database.
Primers sequence specific to different antimicrobials resistant determinants in Salmonella.
| Antibiotic | Gene | Sequence (5′–3′) | Annealing temp (°C) | Amplicon size (bp) |
|---|---|---|---|---|
| Quinolone | gyrA | F-AAATCTGCCCGTGTCGTTGGT | 55 | 343 |
| R-GCCATACCTACGGCGATACC | ||||
| parC | F-CTATGCGATGTCAGAGCTGG | 62 | 270 | |
| R-TAACAGCAGCTCGGCGTATT | ||||
| Tetracycline | tetA | F-GATATTCTGAGCACTGTCGC | 57.5 | 950 |
| R-CTGCCTGGACAACATTGCTT | ||||
| tetB | F-TTGGTTAGGGGCAAGTTTTG | 55 | 600 | |
| R-GTAATGGGCCAATAACACCG | ||||
| tetG | F-GCTCGGTGGTATCTCTGC | 55 | 500 | |
| R-AGCAACAGAATCGGGAAC | ||||
| β-Lactams | Carb | F-AATGGCAATCAGCGCTTCCC | 55 | 586 |
| R-GGGGCTTGATGCTCACTCCA | ||||
| tem | F-TTGGGTGCACGAGTGGGTTA | 55 | 503 | |
| R-GACAGTTACCAATGCTTAATCA | ||||
| oxa-1 | F-ACCAGATTCAACTTTCAA | 55 | 598 | |
| R-TCTTGGCTTTTATGCTTG | ||||
| Chloramphenicol | floR | F-CACGTTGAGCCTCTATAT | 55 | 868 |
| R-ATGCAGAAGTAGAACGCG | ||||
| Trimethoprim | dfrA1 | F-GTGAAACTATCACTAATGG | 50 | 474 |
| R-TTAACCCTTTTGCCAGATTT | ||||
| dfrB | F-GATCACGTGCGCAAGAAATC | 60 | 141 | |
| R-AAGCGCAGCCACAGGATAAAT | ||||
| dfrA14 | F-GAGCAGCTICTITTIAAAGC | 58 | 393 | |
| R-TTAGCCCTTTIICCAATTTT | ||||
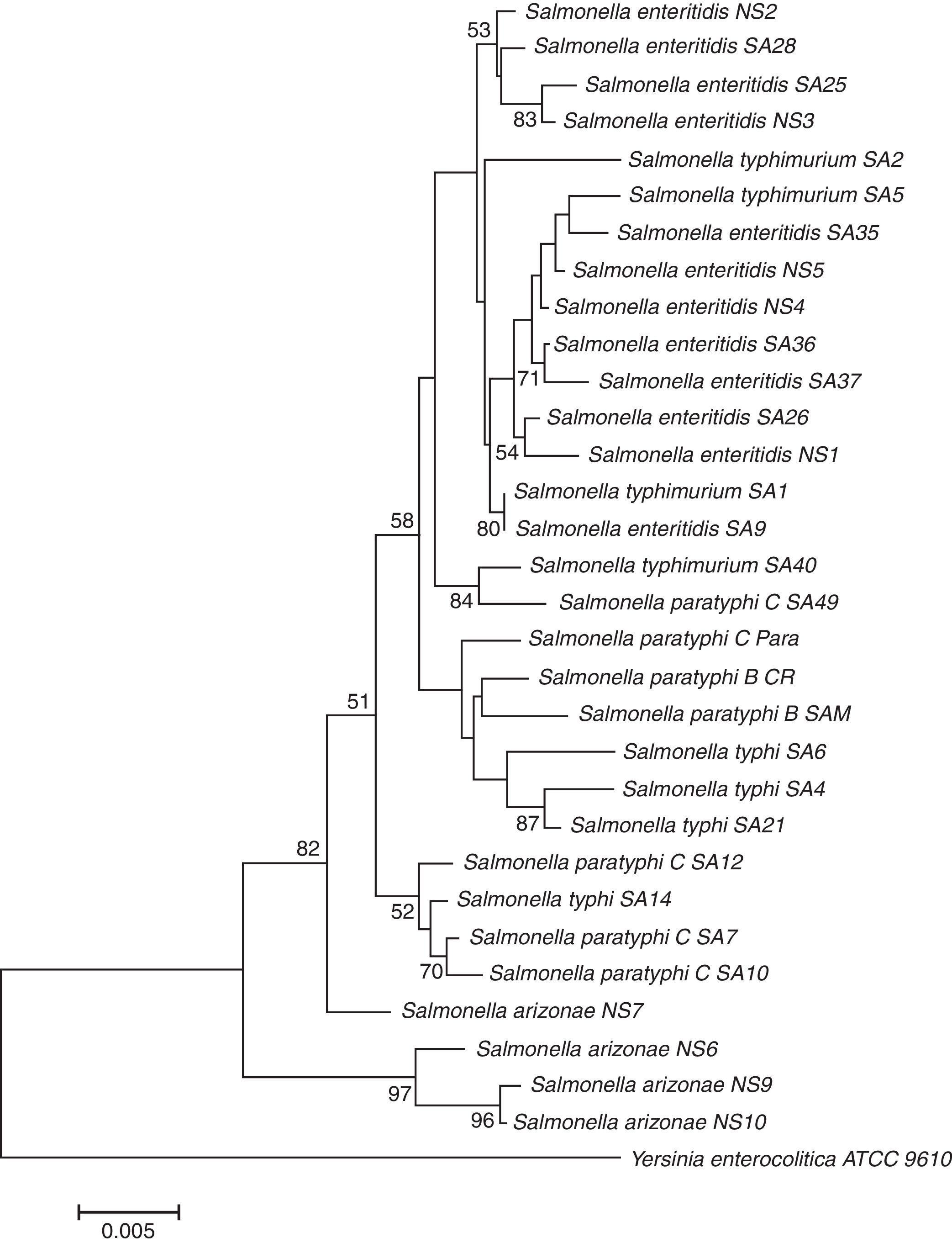
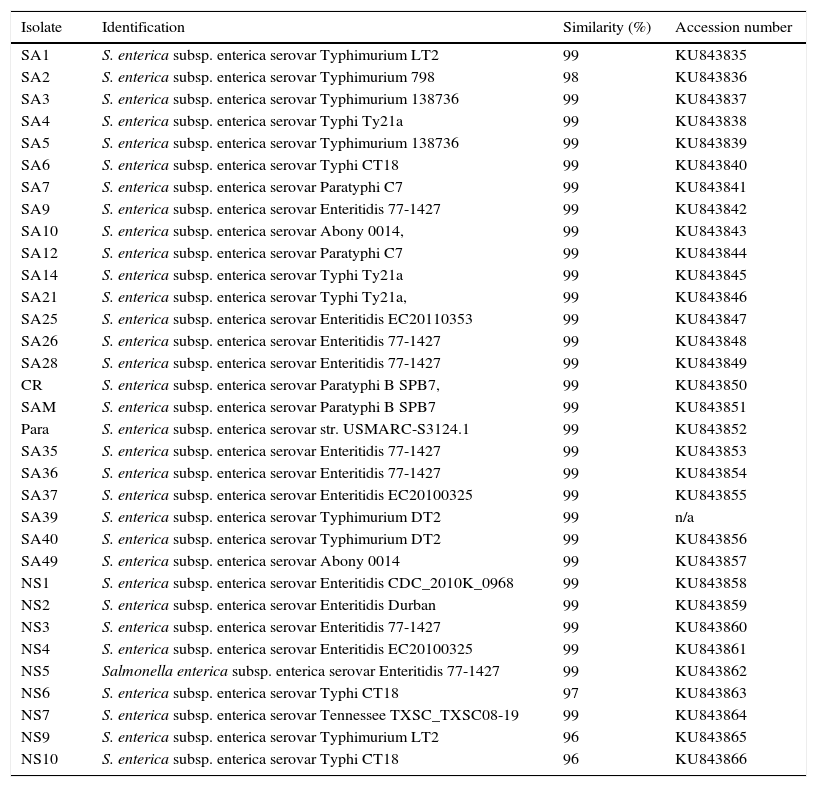
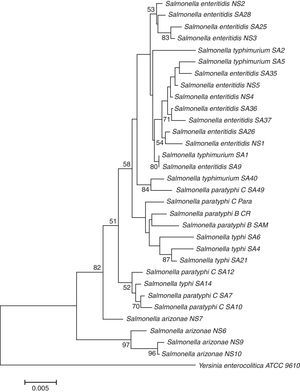
Enrichment and isolation of Salmonella spp. from the collected clinical and environmental samples resulted in isolation of 100 non-repetitive bacterial strains. Among the isolates, 33 strains were identified as Salmonella enterica based on their metabolic reactions and biochemical characteristics. The isolates (n=33) were oxidase negative, produce H2S, and able to utilize arginine, lysine, ornithine, citrate (except one isolate), glucose, mannitol, inositol (variable), sorbitol, rhamnose, melibiose, and arabinose. In addition, all isolates were negative with orth-nitro phenyl-β-d-galactopyranoside (ONPG), tryptophan, urea, indole, Voges Proskauer, gelatin, sucrose, and amygdalin tests. In addition to biochemical tests, the identities of the isolates were further confirmed by 16S rDNA genes sequencing analysis. The 16S rDNA genes of different isolates (n=33) were successfully amplified, with expected length of about 1525bp, purified and sequenced. As shown in Table 2, all isolates (n=33) were affiliated to various strains of Salmonella enterica subsp. enterica with 96–99% similarities; and the sequences were deposited in the GenBank with accession numbers of KU843835 to KU843866. The phylogenetic tree showing the genetic relatedness among the isolated S. enterica strains based on 16S rDNA sequences is shown in Fig. 1.
Identification of Salmonella strains (n=33) based on 16S-rDNA sequencing.
| Isolate | Identification | Similarity (%) | Accession number |
|---|---|---|---|
| SA1 | S. enterica subsp. enterica serovar Typhimurium LT2 | 99 | KU843835 |
| SA2 | S. enterica subsp. enterica serovar Typhimurium 798 | 98 | KU843836 |
| SA3 | S. enterica subsp. enterica serovar Typhimurium 138736 | 99 | KU843837 |
| SA4 | S. enterica subsp. enterica serovar Typhi Ty21a | 99 | KU843838 |
| SA5 | S. enterica subsp. enterica serovar Typhimurium 138736 | 99 | KU843839 |
| SA6 | S. enterica subsp. enterica serovar Typhi CT18 | 99 | KU843840 |
| SA7 | S. enterica subsp. enterica serovar Paratyphi C7 | 99 | KU843841 |
| SA9 | S. enterica subsp. enterica serovar Enteritidis 77-1427 | 99 | KU843842 |
| SA10 | S. enterica subsp. enterica serovar Abony 0014, | 99 | KU843843 |
| SA12 | S. enterica subsp. enterica serovar Paratyphi C7 | 99 | KU843844 |
| SA14 | S. enterica subsp. enterica serovar Typhi Ty21a | 99 | KU843845 |
| SA21 | S. enterica subsp. enterica serovar Typhi Ty21a, | 99 | KU843846 |
| SA25 | S. enterica subsp. enterica serovar Enteritidis EC20110353 | 99 | KU843847 |
| SA26 | S. enterica subsp. enterica serovar Enteritidis 77-1427 | 99 | KU843848 |
| SA28 | S. enterica subsp. enterica serovar Enteritidis 77-1427 | 99 | KU843849 |
| CR | S. enterica subsp. enterica serovar Paratyphi B SPB7, | 99 | KU843850 |
| SAM | S. enterica subsp. enterica serovar Paratyphi B SPB7 | 99 | KU843851 |
| Para | S. enterica subsp. enterica serovar str. USMARC-S3124.1 | 99 | KU843852 |
| SA35 | S. enterica subsp. enterica serovar Enteritidis 77-1427 | 99 | KU843853 |
| SA36 | S. enterica subsp. enterica serovar Enteritidis 77-1427 | 99 | KU843854 |
| SA37 | S. enterica subsp. enterica serovar Enteritidis EC20100325 | 99 | KU843855 |
| SA39 | S. enterica subsp. enterica serovar Typhimurium DT2 | 99 | n/a |
| SA40 | S. enterica subsp. enterica serovar Typhimurium DT2 | 99 | KU843856 |
| SA49 | S. enterica subsp. enterica serovar Abony 0014 | 99 | KU843857 |
| NS1 | S. enterica subsp. enterica serovar Enteritidis CDC_2010K_0968 | 99 | KU843858 |
| NS2 | S. enterica subsp. enterica serovar Enteritidis Durban | 99 | KU843859 |
| NS3 | S. enterica subsp. enterica serovar Enteritidis 77-1427 | 99 | KU843860 |
| NS4 | S. enterica subsp. enterica serovar Enteritidis EC20100325 | 99 | KU843861 |
| NS5 | Salmonella enterica subsp. enterica serovar Enteritidis 77-1427 | 99 | KU843862 |
| NS6 | S. enterica subsp. enterica serovar Typhi CT18 | 97 | KU843863 |
| NS7 | S. enterica subsp. enterica serovar Tennessee TXSC_TXSC08-19 | 99 | KU843864 |
| NS9 | S. enterica subsp. enterica serovar Typhimurium LT2 | 96 | KU843865 |
| NS10 | S. enterica subsp. enterica serovar Typhi CT18 | 96 | KU843866 |
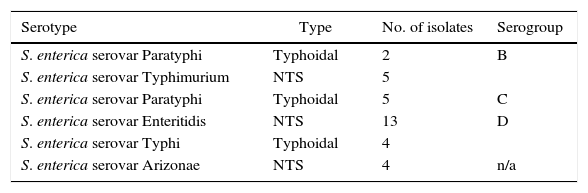
As shown in Table 3, the serotyping of the S. enterica isolates (n=33) revealed that the strains were affiliated to S. enterica Paratyphi B (n=2), S. enterica Typhimurium (n=5), S. enterica Paratyphi C (n=5), S. enterica Enteritidis (n=13), S. enterica Typhi (n=4) and S. enterica Arizonae (n=4). In addition, the serogrouping of S. enterica strains (n=33) indicated 51.52%, 21.21%, and 15.15% were classified as serogroups D, B, and C, respectively. No strains were affiliated to serogroups A or E. In addition, four isolates (12.12%) gave unserotypable isolates using the applied antisera. However, these isolates were identified as S. enterica serovar Arizonae by VITEK 2-C15 identification system.
Serotyping and serogrouping of the isolated S. enterica strains (n=33).
| Serotype | Type | No. of isolates | Serogroup |
|---|---|---|---|
| S. enterica serovar Paratyphi | Typhoidal | 2 | B |
| S. enterica serovar Typhimurium | NTS | 5 | |
| S. enterica serovar Paratyphi | Typhoidal | 5 | C |
| S. enterica serovar Enteritidis | NTS | 13 | D |
| S. enterica serovar Typhi | Typhoidal | 4 | |
| S. enterica serovar Arizonae | NTS | 4 | n/a |
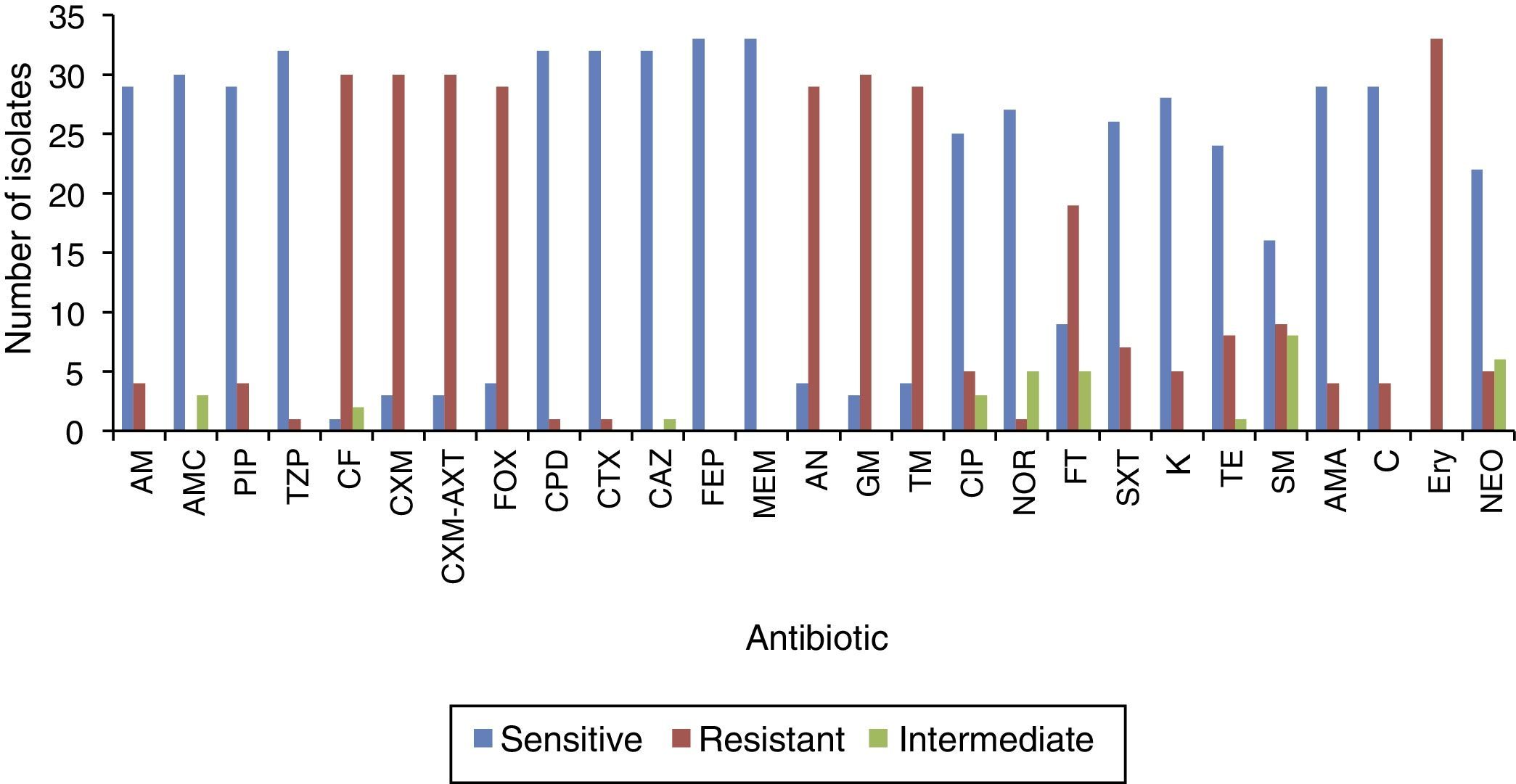
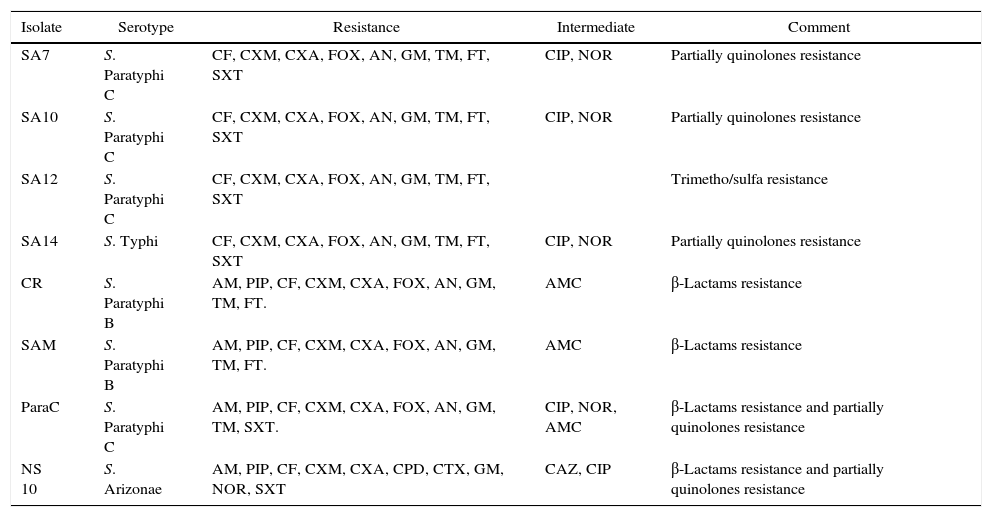
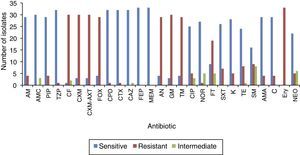
Antibiogram of the isolated S. enterica strains (n=33) to 26 antimicrobial agents including the commonly used antibiotics for salmonellosis treatment is shown in Fig. 2. It was found that among the isolates (n=33), 26 S. enterica strains exhibited multidrug resistance, showing resistance to more than five antibiotics. The results revealed that the highest resistance pattern among all isolates was found to erythromycin (100%), followed by first and second generation of cephalosporin including cefalotin, cefuroxime, cefuroxime axetil (all 90.9%) and cefoxitin (87.9%); and aminoglycosides antibiotics including: gentamicin (90.9%), amikacin and tobramycin (87.9%), respectively. In addition, 57.6% (n=19) of the isolates were resistant to nitrofurantoin, 27.3% (n=9) to streptomycin, 24.2% (n=8) tetracycline, 18.2% (n=6) to trimethoprim–sulfamethoxazole, and 15.2% (n=5) of the isolates were resistant to neomycin. On the other hand, lowest resistance of the S. enterica isolates (n=33) was detected towered piperacillin/tazobactam, cefpodoxime, cefotaxime, and norfloxacin, that only 3.1% of the total isolates exhibited resistance to these antibiotics. In addition, most S. enterica isolates were susceptible to several β-lactams antibiotics (ampicillin, ampicillin subaclam and piperacillin) and chloramphenicol. Noteworthy, the highest level of resistance against most antibiotics was shown among S. Paratyphi C serotype which exhibited high resistance to erythromycin (100% of the isolates), streptomycin (80%), trimethoprim–sulfamethoxazole (80%) tetracycline (80%), neomycin (60%), and kanamycin (60%), followed by S. Paratyphi B which exhibited 100% resistance toward erythromycin, tetracycline, streptomycin, ampicillin-subaclam, and chloramphenicol. Regarding S. Typhi serotype the high level of resistance was shown toward erythromycin (100%), streptomycin (50%), and tetracycline (25%). In addition, five and three isolates exhibited resistance and decreased susceptibility to ciprofloxacin (fluoroquinolone), respectively. Finally, among S. Arizonea isolates (n=4), only one isolate was resistant to kanamycin, tetracycline, trimethoprim–sulfamethoxazole, streptomycin, and ampicillin-subaclam (Table 4).
Frequency of multidrug-resistance patterns among Salmonella enterica isolates (n=33).
| Isolate | Serotype | Resistance | Intermediate | Comment |
|---|---|---|---|---|
| SA7 | S. Paratyphi C | CF, CXM, CXA, FOX, AN, GM, TM, FT, SXT | CIP, NOR | Partially quinolones resistance |
| SA10 | S. Paratyphi C | CF, CXM, CXA, FOX, AN, GM, TM, FT, SXT | CIP, NOR | Partially quinolones resistance |
| SA12 | S. Paratyphi C | CF, CXM, CXA, FOX, AN, GM, TM, FT, SXT | Trimetho/sulfa resistance | |
| SA14 | S. Typhi | CF, CXM, CXA, FOX, AN, GM, TM, FT, SXT | CIP, NOR | Partially quinolones resistance |
| CR | S. Paratyphi B | AM, PIP, CF, CXM, CXA, FOX, AN, GM, TM, FT. | AMC | β-Lactams resistance |
| SAM | S. Paratyphi B | AM, PIP, CF, CXM, CXA, FOX, AN, GM, TM, FT. | AMC | β-Lactams resistance |
| ParaC | S. Paratyphi C | AM, PIP, CF, CXM, CXA, FOX, AN, GM, TM, SXT. | CIP, NOR, AMC | β-Lactams resistance and partially quinolones resistance |
| NS 10 | S. Arizonae | AM, PIP, CF, CXM, CXA, CPD, CTX, GM, NOR, SXT | CAZ, CIP | β-Lactams resistance and partially quinolones resistance |
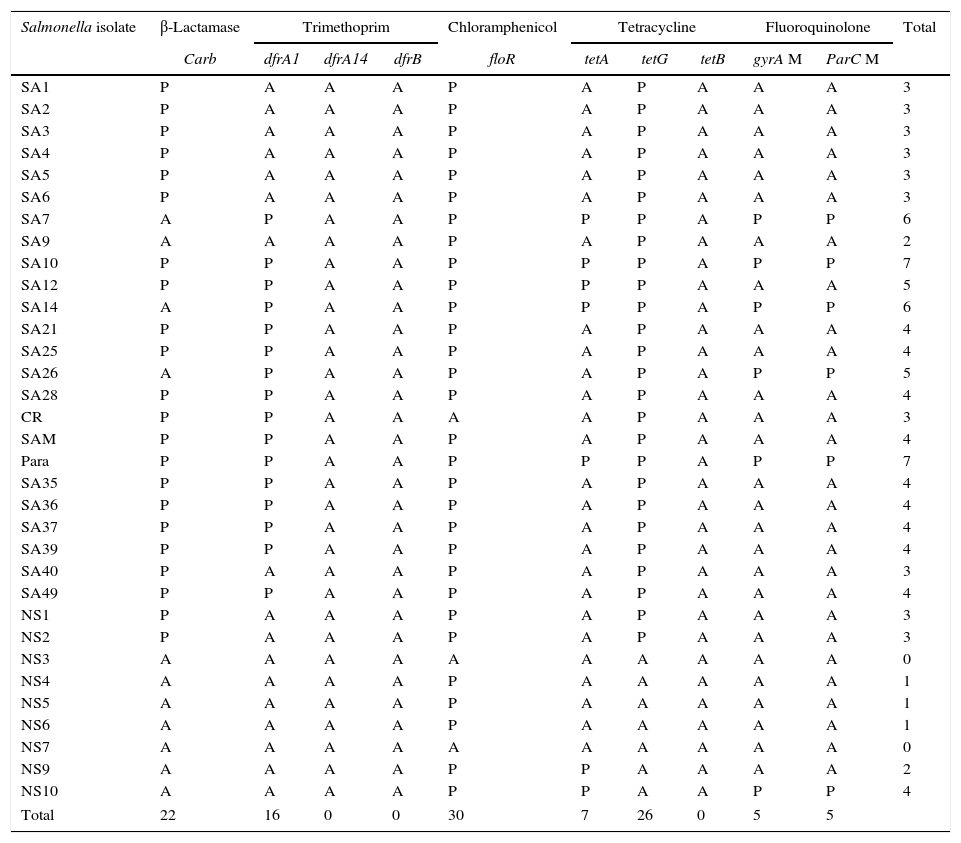
The isolated S. enterica strains (n=33) were screened for the presence of some antibiotic resistance genes by PCR including: tetA, tetB and tetG for tetracycline, carb-like, tem-like and oxa-1-like for β-lactam antibiotics, floR gene for chloramphenicol, dfrA1, dfrB and dfrA14 genes for trimethoprim, and detection of mutation in gyrA (gyrase subunit A) and parC (topoisomerase IV) genes for quinolone residence (Supplementary data). The results summarized in Table 5 revealed the detection of significant variety of resistance determinants among the isolates. Phenotypically, 87.9–90.9% of isolates were resistant to 1st and 2nd generation of cephalosporin, in addition; only four isolates showed ampicillin and piperacillin resistance, and three isolates gave intermediate pattern to amoxicillin/clavulanic acid. The resistance to those β-lactams antibiotics was attributed mainly to presence of carb-like gene which was detected in 22 S. enterica isolates, whereas both tem and oxa-1 genes were absent in all isolates. Tetracycline resistance was related mainly to the presence of the tetA gene, detected in most of the tetracycline resistant isolates (7/8), whereas tetG gene was detected in most isolates. However, tetB gene was absent in all isolates. Despite the presence of floR gene, which confers resistance to chloramphenicol in the most isolates, only four isolates were resistant to chloramphenicol. On the other hand, while all trimethoprim–sulfamethoxazole resistant isolates (n=8) harbored dfrA1 gene, both dfrB and dfrA14 genes were not detected in any isolates, indicating that the resistance to trimethoprim–sulfamethoxazole is mediated by dfrA1 gene. Among the isolated S. enterica strains (n=33), only five isolates exhibited fluoroquinolone resistance. Three of them belonged to S. enterica serovar Paratyphi C (isolates SA7, SA10, and Para), one S. enterica serovar Typhi (SA14) and one S. enterica serovar Arizonae (NS 10). Therefore, the amplified gyrA and ParC genes of those five isolate were purified, sequenced, and the obtained sequences were aligned with reference gyrA and ParC sequences. The results revealed the presence of point mutations in gyrA genes at positions 13 and 24 nucleotides, whereas among parC genes point mutations were detected at positions 13, 19 and 28 nucleotides (Supplementary data).
Distribution of various antibiotic resistance genes (n=11) in S. enterica strains (n=33). P: present; A: absent.
| Salmonella isolate | β-Lactamase | Trimethoprim | Chloramphenicol | Tetracycline | Fluoroquinolone | Total | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Carb | dfrA1 | dfrA14 | dfrB | floR | tetA | tetG | tetB | gyrA M | ParC M | ||
| SA1 | P | A | A | A | P | A | P | A | A | A | 3 |
| SA2 | P | A | A | A | P | A | P | A | A | A | 3 |
| SA3 | P | A | A | A | P | A | P | A | A | A | 3 |
| SA4 | P | A | A | A | P | A | P | A | A | A | 3 |
| SA5 | P | A | A | A | P | A | P | A | A | A | 3 |
| SA6 | P | A | A | A | P | A | P | A | A | A | 3 |
| SA7 | A | P | A | A | P | P | P | A | P | P | 6 |
| SA9 | A | A | A | A | P | A | P | A | A | A | 2 |
| SA10 | P | P | A | A | P | P | P | A | P | P | 7 |
| SA12 | P | P | A | A | P | P | P | A | A | A | 5 |
| SA14 | A | P | A | A | P | P | P | A | P | P | 6 |
| SA21 | P | P | A | A | P | A | P | A | A | A | 4 |
| SA25 | P | P | A | A | P | A | P | A | A | A | 4 |
| SA26 | A | P | A | A | P | A | P | A | P | P | 5 |
| SA28 | P | P | A | A | P | A | P | A | A | A | 4 |
| CR | P | P | A | A | A | A | P | A | A | A | 3 |
| SAM | P | P | A | A | P | A | P | A | A | A | 4 |
| Para | P | P | A | A | P | P | P | A | P | P | 7 |
| SA35 | P | P | A | A | P | A | P | A | A | A | 4 |
| SA36 | P | P | A | A | P | A | P | A | A | A | 4 |
| SA37 | P | P | A | A | P | A | P | A | A | A | 4 |
| SA39 | P | P | A | A | P | A | P | A | A | A | 4 |
| SA40 | P | A | A | A | P | A | P | A | A | A | 3 |
| SA49 | P | P | A | A | P | A | P | A | A | A | 4 |
| NS1 | P | A | A | A | P | A | P | A | A | A | 3 |
| NS2 | P | A | A | A | P | A | P | A | A | A | 3 |
| NS3 | A | A | A | A | A | A | A | A | A | A | 0 |
| NS4 | A | A | A | A | P | A | A | A | A | A | 1 |
| NS5 | A | A | A | A | P | A | A | A | A | A | 1 |
| NS6 | A | A | A | A | P | A | A | A | A | A | 1 |
| NS7 | A | A | A | A | A | A | A | A | A | A | 0 |
| NS9 | A | A | A | A | P | P | A | A | A | A | 2 |
| NS10 | A | A | A | A | P | P | A | A | P | P | 4 |
| Total | 22 | 16 | 0 | 0 | 30 | 7 | 26 | 0 | 5 | 5 | |
Salmonellosis is considered as an immense public health challenge with a reported increase in its incidence.15 The emergence of multidrug resistant Salmonella strains represents a big health challenge and can lead to more acute and invasive infections, in addition to treatment failures owing to resistance would increase the risk of mortality, particularly in the developing countries.16,17Salmonella spp. is one of the most important pathogen that causes food poisoning in Saudi Arabia, particularly in Umrah and Hajj seasons that a lot of tourists are visiting the holy places in Saudi Arabia.18 In this study 33 clinical and environmental bacterial strains were isolated and identified as S. enterica based on their biochemical characterizations and 16S rDNA genes sequences analysis. It was found that the prevalence of non-typhoidal Salmonella (n=18) is more frequent than the typhoid one (n=11). S. Enteritidis and S. Typhimurium represented 39.4% and 15.2% of the total isolates (n=33) respectively, whereas typhoidal Salmonella including S. Paratyphi and S. Typhi represented 21.2% and 12.1% respectively. In addition, 12.1% of the isolates belonged to S. Arizonae. These results were in accordance with various global studies, where S. Enteritidis was the most dominant serotype among the isolated S. enterica strains.18–21 However, in a recent study in Belgium reported by Ceyssens et al.,5 the dominant serotype was S. enterica Typhimurium (55%) followed by Enteritidis (19%).
Investigation of susceptibility of the isolated S. enterica strains (n=33) toward various antibiotics (n=26), indicated that there was high level of antibiotics resistance among isolated S. enterica strains, 26 isolates exhibited multidrug resistance, showing resistance to more than three unrelated antibiotics. Regarding β-lactams antibiotics, 20% of S. Paratyphi C isolates were resistant to the first-line antibiotics, ampicillin-subaclam and chloramphenicol, 100% of S. Paratyphi B were resistant to those two antibiotics. In addition, Paratyphi C isolates showed high resistance to erythromycin, tetracycline, neomycin and kanamycin. This resistance pattern were in agreement with several previous reports.22,23 However, in contrast to several studies which reported high resistance of S. Typhi strains to all first-line drugs, our results revealed a highest susceptibility among S. Typhi isolates (80%) to both ampicillin-subaclam, and chloramphenicol.24,25 In addition, all isolates (n=33/33) and most isolates (n=24/33) exhibited resistance to erythromycin and nitrofurans, respectively. The high resistance of Salmonella to those antibiotics is likely due to the veterinary use of nitrofurans and erythromycin as feed supplement and/or treatment; particularly poultry sector.26–28 Moreover, the results revealed the emergence of two isolate (6.1%) showing resistance to third-generation cephalosporin antibiotics (Cefpodoxime and Cefotaxime), which is less than a study carried out by Burke et al7 who reported that 11% of the S. enterica isolates exhibiting resistance to third-generation cephalosporin. Among the isolated S. enterica isolates (n=33), five and three isolates showed resistance and decreased susceptibility to ciprofloxacin (quinolone), respectively. However, emergence of higher quinolone resistance among S. enterica strains to quinolone has been reported.5
Analysis of resistance determinants in the isolated S. enterica strains (n=33) revealed the detection of carb-like gene (carbenicillinase) in the isolates that exhibited resistance or decreased susceptibility to β-lactam antibiotics, suggesting that this resistance is mediated by carb-like gene which encoded β-lactamase enzyme. Both tem and oxa-1 genes could not be detected in any isolate which is in contrast to other studies where ampicillin-resistance in S. enterica isolates were attributed to blaTEM-1 and blaoxa-1.5,17,23 It was found that the five isolates that exhibited resistance to trimethoprim–sulfamethoxazole were associated with presence of dfrA1 gene (dfrA14 and dfrB were not detected in any isolate), indicating it is responsible for the resistance. However, dfrA1 was not found in resistant S. Arizonae isolate, suggesting that the resistance trimethoprim–sulfamethoxazole in S. Arizonae is attributed to other mechanisms. It was reported that dfrA1 is the most prevalence in S. enterica isolates from Europe, whereas the most common dfrA genes in Korea and Australia are dfrA17 and dfrA12, respectively.23,29–31 The resistance to chloramphenicol is highly associated with the acquisition and expression of efflux pumps that reduce toxic levels of the drug in the bacterial cells. In Salmonella, chloramphenicol efflux pumps are encoded by floR or cml.27floR gene was detected in most tested isolates (n=33). However, only four S. enterica isolates exhibited resistance to chloramphenicol. This finding is supported by other studies that reported the presence of floR gene in various S. enterica as part of Salmonella pathogenicity island-1.23,32 The resistance to tetracycline is highly associated with the acquisition and expression of efflux pumps, encoded by tet genes, that reduce the concentration of the drug inside the bacterial cells. Out of eight of isolates exhibited resistance patterns to tetracycline, seven isolates harbored tetA gene. This result was in agreement with the hypothesis said the intestinal tract is a suitable niche for the transfer of tetA and tetB by horizontal gene transfer thereby these genes are popular among Enterobacteriaceae.33 Quinolones resistance are usually mediated mainly by point mutations in bacterial gyrase (gyrA and gyrB) and topoisomerase IV (parC and parE) genes. These mutations lead to block the binding site of topoisomerase or gyrase targeting by antimicrobial agents.34,35 In the present study point mutations were detected in all quinolones resistant S. enterica isolates (n=5) in both gyrA and parC with large changes in both Para and NS10 isolates leading to complete frame shift of amino acids sequences of proteins in both topoisomerase and gyrase. Substitutions among SA7, SA10, SA14 isolates occurred in gyrA gene in both position 13 and 24 of nucleotides, which led to single amino acid substitution (serine instead of Phenylalanine) in the three isolates while aspartate was replaced by tyrosine in both S. Paratyphi C isolates. On the other hand, a high variation was detected in parC gene among the resistant isolates causing major changes in their proteins. The presence of these mutations in both parC and gyrA renders these isolates to be more resistant to fluoroquinolones. Similar results of point mutations in both parC and gyrA genes were reported, one mutation in gyrA (Asp87Asn) and one in parC (Thr54Ser)17; and point mutations in GyrA residues Ser83 and Asp87 and ParC_Ser80Ile,5 that conferred quinolones resistance in S. enterica.
ConclusionIn this study, we report epidemiology, antimicrobial susceptibility, and the genetic basis of resistance among S. enterica strains isolated in Saudi Arabia. The obtained results alarm the emergence of MDR Salmonella enterica isolated in Saudi Arabia, showing resistance to first line drug as well as to third generation cephalosporin in Saudi Arabia. In addition, it describe some details about the molecular mechanism of the resistance which revealed and support the hypothesis that the antimicrobial resistance mechanism in S. enterica is varied according to the geographic area and based on the environment of isolation. The obtained data is a basis for further investigation on large scale samples for more understating of the Salmonellosis in Saudi Arabia.
Conflict of interestThe authors have no potential conflict of interest.
The authors extend their appreciation to the Research Center at College of Science, Deanship of Scientific Research at King Saud University for funding this work.