Vitellibacter aquimaris D-24T (=KCTC 42708T=DSM 101732T), a halophilic marine bacterium, was isolated from seawater collected from Desaru beach, Malaysia. Here, we present the draft genome sequence of D-24T with a genome size of approximately 3.1Mbp and G+C content of 39.93%. The genome of D-24T contains genes involved in reducing a potent greenhouse gas (N2O) in the environment and the degradation of proteinaceous compounds. Genome availability will provide insights into potential biotechnological and environmental applications of this bacterium.
Over the last few decades, marine microorganisms have received considerable attention for their applications in biotechnology. Prime focus has been given on identification of halophilic enzymes with potential applications.1,2 Halophilic enzymes boast unique properties such as the sustainability at extreme ranges of salt concentration, temperature, pH, and organic solvents.3,4 Characterization of marine bacteria through their genome sequence has provided great opportunities for mining and identifying of enzymes with biotechnological importance.
Vitellibacter aquimaris D-24T, previously isolated from seawater, was confirmed as a new species in genus Vitellibacter.5 This bacterium is a Gram negative, rod-shape and yellow-orange pigmented bacterium. In this study, we report the draft genome of V. aquimaris D-24T.
V. aquimaris D-24T was grown in Marine Broth 2216 (BD Difco) and the genomic DNA was extracted using the DNeasy Blood and Tissue kit (Qiagen, Hilden, Germany) per manufacturer instructions. The genome of the V. aquimaris D-24T sequence was generated using pair-end sequencing in an Illumina MiSeq sequencing platform (Illumina, California, USA). De novo assembly was performed using IDBA-UD version 1.0.9,6 obtaining 66 contigs with a N50 contig length of 439,587. Genome annotations were carried out using NCBI Prokaryotic Genome Annotation Pipeline 2.87 and Integrated Microbial Genomes Expert Review (IMG-ER).8 Protein coding sequences were predicted by using GeneMarkS+ version 2.8 with the best-placed reference protein method9 and Prodigal,10 respectively.
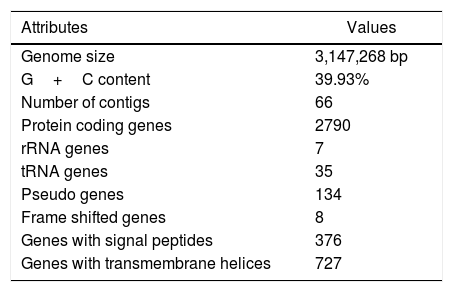
The draft genome of V. aquimaris D-24T is comprised of 3,147,268 nucleotides with a mean G+C content of 39.93%. A total of 2967 genes were predicted, of which 2790 (94.03%) protein-coding genes were assigned with putative function or hypothetical proteins. The genome consists of three 5S rRNA, one 16S rRNA, one 23 rRNA, and 35 tRNA genes (Table 1). The genome V. aquimaris D-24T contains genes encoded for nitrite reductase (nirK) (WP_045079976.1), nitric oxide reductase subunit B (norB) (WP_062619270.1), nitric oxide reductase subunit C (norC) (WP_062619272.1), and nitrous oxide reductase (nosZ) (WP_062619280.1), all of which are involved in the denitrification pathway (NO2− to N2). Nitrous oxide (N2O) is a potent greenhouse gas which also damages the Earth's ozone layer. Due to intensive agricultural and industrial practices, the release of this gas into the atmosphere has greatly increased.11,12 Nitrous-oxide reductase plays an important role in catalyzing the reduction of nitrous oxide to nitrogen (N2).13 The nitrous oxide reductase (nosZ) from V. aquimaris D-24T is a promising candidate for removing nitrous oxide, a potent greenhouse gas, from the environment.
Genome features of Vitellibacter aquimaris D-24T.
| Attributes | Values |
|---|---|
| Genome size | 3,147,268 bp |
| G+C content | 39.93% |
| Number of contigs | 66 |
| Protein coding genes | 2790 |
| rRNA genes | 7 |
| tRNA genes | 35 |
| Pseudo genes | 134 |
| Frame shifted genes | 8 |
| Genes with signal peptides | 376 |
| Genes with transmembrane helices | 727 |
Genome analysis also revealed a total of twelve genes encoded for proteases, including two metalloproteases (KXO00999.1; KXN98592.1); two serine proteases (KXO01375.1; KXN97930.1); five proteases (KXO00989.1; KXO00819.1; KXN99950.1; KXN99951.1; KXO00100.1); two zinc metalloproteases (KXO00568.1; KXN98177.1); and an acid protease (KXO00796.1). Genes encoding proteases were also present in the genome of another species of Vitellibacter.14 The draft genome sequence of V. aquimaris D-24T will facilitate understanding and development of potential applications of Vitellibacter species.
The Whole Genome Shotgun project has been deposited at DDBJ/EMBL/GenBank under the accession number JRWG00000000. The first version (accession number JRWG01000000) is described in this paper.
Conflicts of interestThe authors declare no conflicts of interest.
This work was financially supported by Ministry of Education Malaysia (grant number: 4F265) and Universiti Teknologi Malaysia RU grants (grant number: 07H43 and 14H67). Suganthi Thevarajoo and Chitra Selvaratnam acknowledge the PhD Zamalah scholarship from Universiti Teknologi Malaysia. Kok-Gan Chan acknowledges the University of Malaya High Impact Research Grants (UM.C/625/1/HIR/MOHE/CHAN/01, Grant No. A-000001-50001 and UM-MOHE HIR Grant UM.C/625/1/HIR/MOHE/CHAN/14/1, no. H-50001-A000027).