As the largest genus in Actinobacteria family, Streptomyces species have the ability to synthesize numerous compounds of diverse structures with bioactivities. Streptomyces mangrovisoli MUSC 149T was previously isolated as a novel streptomycete from mangrove forest in east coast of Peninsular Malaysia. The high quality draft genome of MUSC 149T comprises 9,165,825bp with G+C content of 72.5%. Through bioinformatics analysis, 21 gene clusters identified in the genome were associated with the production of bioactive secondary metabolites. The presence of these biosynthetic gene clusters in MUSC 149T suggests the potential exploitation of the strain for production of medically important compounds.
Members of Streptomyces genus have received considerable attention and sparked interest among pharmaceutical industry as they are capable of synthesizing compounds of diverse structures with bioactivities including antibiotics, anti-rejection (immunosuppressant), antioxidant and anticancer.1–5Streptomyces mangrovisoli MUSC 149T was previously isolated as a novel streptomycete with antioxidant potential from mangrove forest in east coast of Peninsular Malaysia6,7 and the strain has been deposited at two culture collection centers (=MCCC 1K00252T=DSM 42140T). Thus, the strain MUSC 149T was selected for genome sequencing as an attempt to identify biosynthetic gene clusters associated with secondary metabolite production.
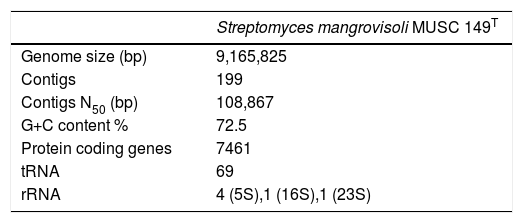
The genomic DNA of MUSC 149T was extracted with Masterpure™ DNA purification kit (Epicentre, Illumina Inc., Madison, WI, USA) followed by RNase (Qiagen, USA) treatment.8,9 DNA quality was examined using NanoDrop spectrophotometer (Thermo Scientific, Waltham, MA, USA) and a Qubit version 2.0 fluorometer (Life Technologies, Carlsbad, CA, USA). Subsequently, DNA library was constructed using Nextera™ DNA Sample Preparation kit (Nextera, USA) and the library quality was validated by Bioanalyzer 2100 high sensitivity DNA kit (Agilent Technologies, Palo Alto, CA) before performing paired-end on MiSeq platform with MiSeq Reagent Kit 2 (2× 250bp; Illumina Inc., Madison, WI, USA). The paired-end reads were trimmed and de novo assembled with CLC Genomics Workbench version 7 (CLC bio, Denmark). The analysis generated 199 contigs with N50 size of 108,867bp (Table 1). The assembled genome size of MUSC 149T contained 9,165,825bp, with an average coverage of 119.0-fold and G+C content of 72.5%. The whole genome project of MUSC 149T was deposited at DDBJ/EMBL/GenBank under accession number LAVA00000000 and the version described in this paper is the second version (LAVA02000000).
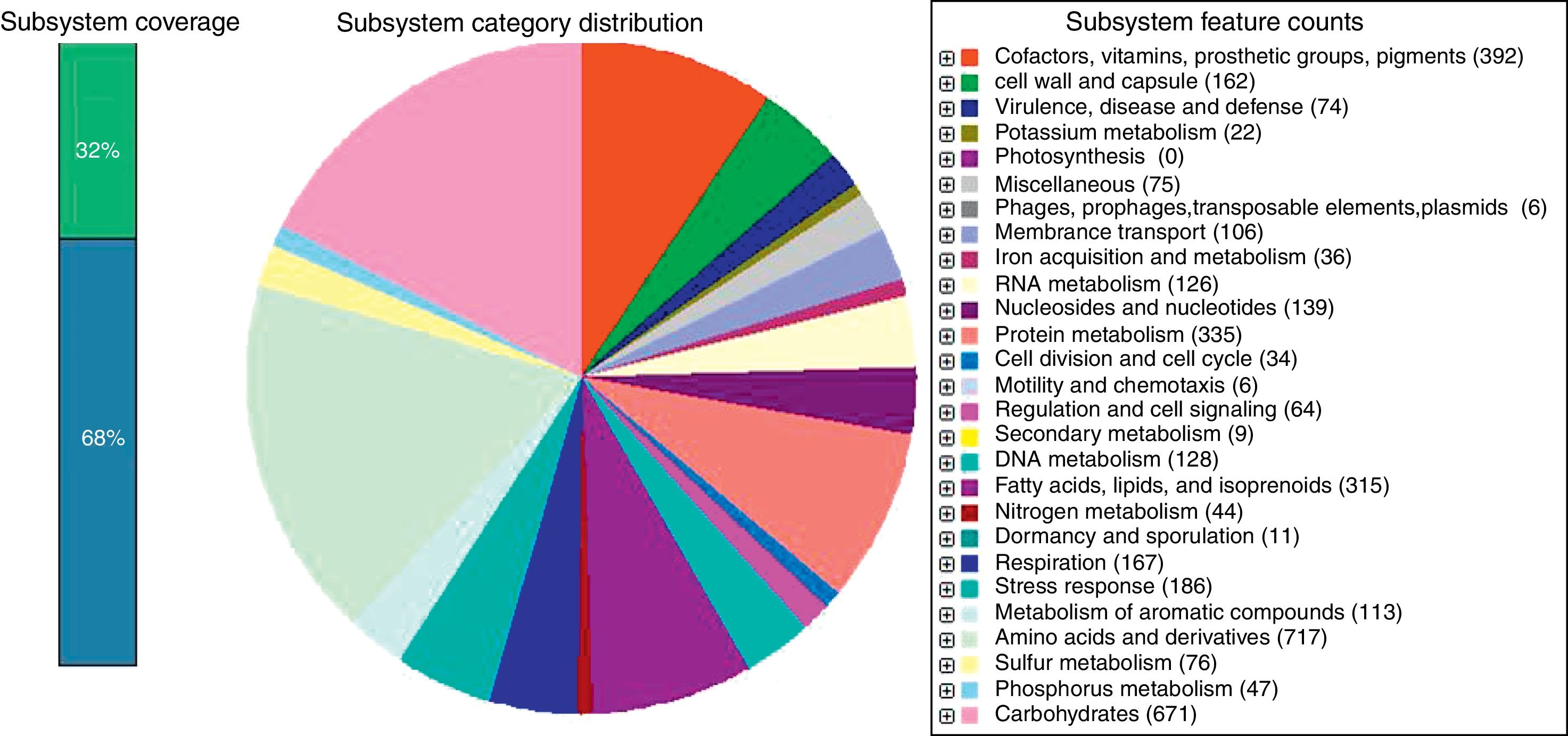
Gene prediction was performed using Prodigal version 2.6, whereas rRNA and tRNA were predicted using RNAmmer and tRNAscan SE version 1.21.10–12 The assembly was uploaded for annotation to Rapid Annotation using Subsystem Technology (RAST).13 A total of 7461 protein-encoding genes was predicted and assigned to 442 subsystems, along with 69 tRNA and 6 rRNA genes (Fig. 1). Among the subsystems, most of the genes were involved in amino acids and derivatives metabolism (8.97%), followed by carbohydrates metabolism (8.40%) and cofactors, vitamins, prosthetic groups, pigments metabolism subsystems (4.91%).
In order to investigate its bioactive potential, the genome of MUSC 149T was further analyzed using antibiotics & Secondary Metabolite analysis shell (antiSMASH) version 3.0.14 The antiSMASH server revealed 21 gene clusters related to antibiotics and secondary metabolite biosynthesis, with four clusters showed more than 70% similarities to known gene clusters: venezuelin biosynthetic gene cluster (75%), ectoine biosynthetic gene cluster (75%), desferrioxamine B biosynthetic gene cluster (83%), and hopene biosynthetic gene cluster (85%). Previously, extract of MUSC 149T was found to possess antioxidant activity; the presence of siderophores biosynthetic gene clusters such as desferrioxamine B has further highlight the antioxidant potential of strain MUSC 149T.
In conclusion, we report the draft genome sequence of S. mangrovisoli MUSC 149T. The availability of its genome sequence has revealed production of potentially medically useful compounds and certainly deserves further detailed study.
Conflicts of interestThe authors declare no conflicts of interest.
This work was supported by PVC Award Grant (Project No. PVC-ECR-2016), External Industry Grant (Biotek Abadi Vote No. GBA-808813), Fundamental Research Grant Scheme (FRGS/1/2013/SKK01/MUSM/03/3), MOSTI eScience funds (Project No. 06-02-10-SF0300) awarded to L.-H.L. and MOSTI eScience funds (Project No. 02-02-10-SF0215) awarded to B.-H.G., and a University of Malaya for High Impact Research Grant (UM-MOHE HIR Nature Microbiome Grant No. H-50001-A000027 and No. A000001-50001) and PPP Grant (PG090-2015B) awarded to K.-G.C.