Hepatitis E virus is responsible for acute and chronic liver infections worldwide. Swine hepatitis E virus has been isolated in Brazil, and a probable zoonotic transmission has been described, although data are still scarce. The aim of this study was to investigate the frequency of hepatitis E virus infection in pigs from a small-scale farm in the rural area of Paraná State, South Brazil. Fecal samples were collected from 170 pigs and screened for hepatitis E virus RNA using a duplex real-time RT-PCR targeting a highly conserved 70nt long sequence within overlapping parts of ORF2 and ORF3 as well as a 113nt sequence of ORF2. Positive samples with high viral loads were subjected to direct sequencing and phylogenetic analysis. hepatitis E virus RNA was detected in 34 (20.0%) of the 170 pigs following positive results in at least one set of screening real-time RT-PCR primers and probes. The swine hepatitis E virus strains clustered with the genotype hepatitis E virus-3b reference sequences in the phylogenetic analysis and showed close similarity to human hepatitis E virus isolates previously reported in Brazil.
In endemic areas, hepatitis E virus (HEV) causes large epidemics and sporadic cases in humans, including genotype HEV-1 in Asia and Africa, genotype HEV-2 in Mexico and Africa, and genotype HEV-4 in Asia. In non-endemic areas, isolated cases of genotype HEV-3 occur in Europe, Japan and the Americas. Genotypes HEV-3 and HEV-4 are described as zoonotic, as they infect numerous mammalian species, including domestic pigs, and can be transmitted through the ingestion of raw or undercooked meat from infected animals.1
HEV shows notable heterogeneity with several groups and genotypes. A recent consensus has classified this virus in one family of Hepeviridae, divided in 2 genera namely Orthohepevirus and Piscihepevirus. Orthohepevirus is further divided into 4 species from A to D. Orthohepevirus A is the species infecting humans and swine and other animals, such as boar, deer, mongoose, rabbit and camel. These include two genotypes isolated from humans alone (HEV-1 and HEV-2), two genotypes reported in both humans and different animal species and associated with the zoonotic cases (HEV-3 and HEV-4), two isolates from wild boar in Japan (genotype HEV-5 and HEV-6) and a single isolate from dromedary camel in Dubai (genotype HEV-7). Rabbit HEV and closely related human isolate have been placed as a distant member in HEV-3. The moose virus appears to cluster closely to genotype HEV-3, while HEV isolates from mink to ferret virus (HEV C2) and HEV isolates from fox to rat virus (HEV C1). These viruses have not been placed in any specific genotype and need complete genomic sequences for a definite taxonomic classification. Orthohepevirus B includes all 3 avian HEV strains, Orthohepevirus C one species from rat and another one from ferret and Orthohepevirus D includes bat HEV. Piscihepevirus includes 2 trout HEV strains within a single species.2,3
In Brazil, swine HEV was first isolated from pig fecal samples from the São Paulo State, Southeastern Brazil.4 Moreover, genotype HEV-3 was found in pigs and effluents from a pig slaughterhouse in Rio de Janeiro State, Southeastern Brazil,5,6 and in pigs from the Eastern Brazilian Amazon7 and from Southern Brazil.8,9 Nonetheless, hepatitis E has only been studied as a potential zoonotic disease in Latin America in the last ten years, and data on this subject are still scarce.
Data on human HEV in Brazil are limited as well. This country has been classified as moderately endemic for HEV, with seroprevalence from 1% to 4% in blood donors or the general population, 13% in individuals from an agricultural settlement in the Amazon Basin, and 15% in renal transplant recipients.10–13 A more recent study observed a seroprevalence of anti-HEV IgG antibodies of 10% among blood donors in the metropolitan area of Itajai Valley, Southern Brazil, a region of predominant German ancestry where cultural habits result in a high pork consumption.14 Genotype HEV-3 infection has been described in the country, both among immunocompetent and immunocompromised individuals.15–17
Brazil is the fourth biggest pork exporter in the world, with an important increase in recent years. The State of Paraná has one of the largest swine productions in the country, and in 2014, this region was solely responsible for 20% of the national pig slaughter. In addition to export, pork consumption has also increased in Brazil.18 Specifically in Paraná State, the predominant European ancestry19,20 leads to higher pork consumption than in other parts of the country.21,22 The impact of such habits and the potential transmission of swine HEV to humans in this region remain unknown.
In the present study, we investigated the frequency of HEV infection in pigs from a small-scale farm in the rural area of Paraná State, South Brazil.
Materials and methodsFecal samplesThe study protocol was approved by the Institutional Committee for Ethics in the Use of Research Animals (CEUA-UNIFESP 2014/1004300914).
In September 2014, fecal samples were collected from 170 pigs from a small-scale farm in the municipality of Itapejara d’Oeste, in a rural area of Paraná State, Southern Brazil. This farm is one of the many small farms in the region, with a current population of approximately 70 sows, 5 boars and 580 pigs that are sold to slaughterhouses when raised to a median age of 22 weeks. The sample size of 170 animals was calculated to allow determining a HEV RNA prevalence of approximately 15%9 with a 95% confidence interval (CI). Additionally, sampling was performed at the ages of 4, 7, 10, 13 and 16 weeks, according to the age distribution of all the pigs raised for slaughter.
RNA extraction, nested RT-PCR and quantitative RT-PCRHEV RNA was extracted from the fecal samples using the QIAamp viral RNA mini kit (QIAGEN, Hilden, Germany). Briefly, fecal samples were first suspended in a 50% solution in ultrapure nuclease-free distilled water, then centrifuged at 20,000×g for 15min, and the supernatant was used to extract viral RNA according to the manufacturer's instructions. Quantitative RT-PCR was performed according to a previously described modified one-step duplex real-time protocol,23 with a primer and probe set targeting a highly conserved 70nt long sequence within overlapping parts of ORF2 and ORF324 as well as a set specific for a 113nt sequence of ORF2.25 A previously characterized plasmid clone from a Brazilian human HEV strain (KF152884)17 was constructed with the TOPO® TA Cloning® Kit (Invitrogen, Carlsbad, CA, USA) and the described primers. Plasmid DNA was purified using the QIAprep Spin Miniprep Kit (Qiagen, Hilden, Germany) and then linearized and quantified with the Nanodrop ND-1000 instrument (Wilmington, DE, USA), followed by transcription to RNA with T7 RNA polymerase (Promega, Madison, WI, USA). Standard curves were generated using 10−1–1010 copies of plasmid RNA. HEV viral loads were determined based on standard curves and were reported as the log10 number of copies of HEV RNA per mL of fecal suspension. The detection limit of the real-time RT-PCR was three copies of viral RNA per reaction (2.40log10 copies per mL of fecal suspension), while the quantification limit was set at 10 copies of viral RNA per reaction (3.00log10 copies per mL of fecal suspension). All screening reactions were run in duplicate with proper controls, whereas positive results were confirmed in separate confirmatory reactions.
The qualitative nested RT-PCR one-step reaction was conducted with primers to amplify partial regions of ORF1 and ORF2 of 287 nt and 348 nt, respectively, after second-round PCR.26,27 All precautions and procedures suggested to avoid the possibility of cross-contamination were employed. Amplified products were visualized in a 1.5% agarose gel stained with SYBR® Safe (Life Technologies, Austin, TX, USA).
Sequencing and phylogenetic analysisFinal fragments obtained from the nested RT-PCR analysis (ORF1 287 nt and ORF2 348nt) were purified using the ExoSAP-IT PCR Clean-up Kit (GE Healthcare, Chalfont St. Giles, UK), and sequenced using the BigDye® Terminator v3.1 Cycle Sequencing Kit and the automated ABI 3100 DNA Sequencer (Applied Biosystems, Foster City, CA, USA), according to the manufacturer's instructions. Sequences from human and swine HEV were collected from public databases, and phylogenetic trees were constructed using the neighbor-joining method with the Kimura 2-parameter model of nucleotide substitution in MEGA v. 5.0 (The Biodesign Institute, USA). Statistics was performed by bootstrap analysis with 1000 pseudoreplicates. The sequences reported in this study are available in the GenBank database under the accession numbers KP966825–KP966829.
Statistical analysisAll data were analyzed using SPSS version 11.0 (SPSS Inc., Chicago, IL, USA). Descriptive statistics consisted of the characterization of the studied population and the frequency of HEV-RNA detection with the respective percentages and 95% CI. The bivariate analysis to compare categorical values consisted of Pearson's Chi-square test. Non-conditional logistic regression was used to identify associations between dependent and independent variables by the means of odds ratio (OR). For this analysis, the ages of 4 and 7 weeks were grouped to avoid empty cells, as well as the ages of 13 and 16 weeks. The statistical significance level was p<0.05. All reported values are two-tailed.
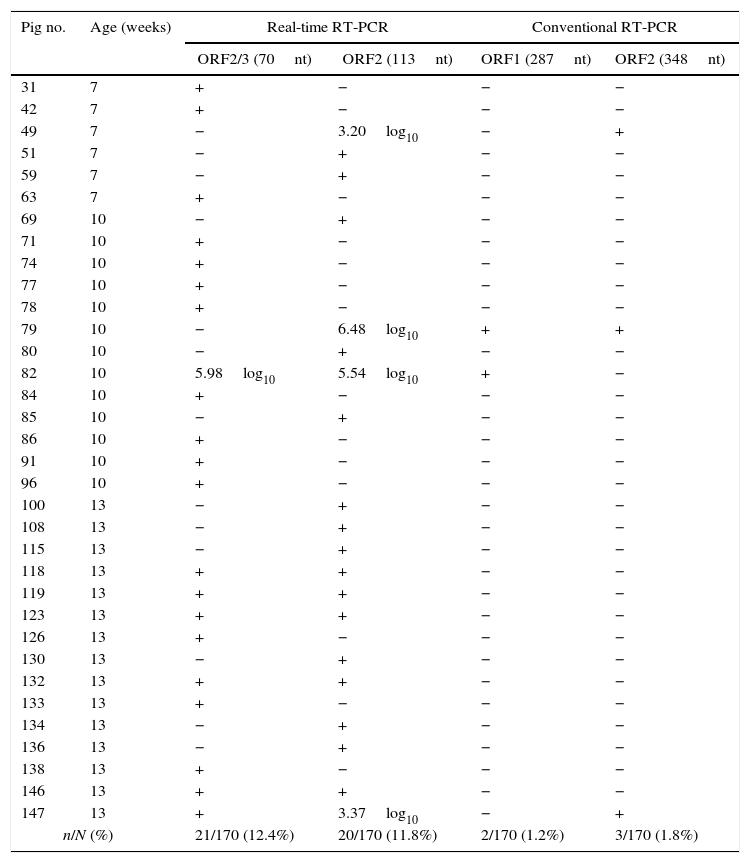
ResultsHEV-RNA detection and sequencingHEV-RNA was detected in 34 (20.0%) of 170 pigs following a positive result in at least one set of screening real-time RT-PCR primers and probes. Seven (20.6%) of these samples were positive with both sets of primers and probes. The 70 nt ORF2/3 set amplified 21 (12.4%) of 170 samples, while the 113 nt ORF2 set amplified 20 samples (11.8%). Among the 34 positive samples, only 4 (11.8%) presented viral loads higher than 3.00log10 copies per mL of fecal suspension (Table 1).
Molecular detection of HEV-RNA in pigs from the rural area of Paraná State, South Brazil, using real-time and conventional RT-PCR.
| Pig no. | Age (weeks) | Real-time RT-PCR | Conventional RT-PCR | ||
|---|---|---|---|---|---|
| ORF2/3 (70nt) | ORF2 (113nt) | ORF1 (287nt) | ORF2 (348nt) | ||
| 31 | 7 | + | − | − | − |
| 42 | 7 | + | − | − | − |
| 49 | 7 | − | 3.20log10 | − | + |
| 51 | 7 | − | + | − | − |
| 59 | 7 | − | + | − | − |
| 63 | 7 | + | − | − | − |
| 69 | 10 | − | + | − | − |
| 71 | 10 | + | − | − | − |
| 74 | 10 | + | − | − | − |
| 77 | 10 | + | − | − | − |
| 78 | 10 | + | − | − | − |
| 79 | 10 | − | 6.48log10 | + | + |
| 80 | 10 | − | + | − | − |
| 82 | 10 | 5.98log10 | 5.54log10 | + | − |
| 84 | 10 | + | − | − | − |
| 85 | 10 | − | + | − | − |
| 86 | 10 | + | − | − | − |
| 91 | 10 | + | − | − | − |
| 96 | 10 | + | − | − | − |
| 100 | 13 | − | + | − | − |
| 108 | 13 | − | + | − | − |
| 115 | 13 | − | + | − | − |
| 118 | 13 | + | + | − | − |
| 119 | 13 | + | + | − | − |
| 123 | 13 | + | + | − | − |
| 126 | 13 | + | − | − | − |
| 130 | 13 | − | + | − | − |
| 132 | 13 | + | + | − | − |
| 133 | 13 | + | − | − | − |
| 134 | 13 | − | + | − | − |
| 136 | 13 | − | + | − | − |
| 138 | 13 | + | − | − | − |
| 146 | 13 | + | + | − | − |
| 147 | 13 | + | 3.37log10 | − | + |
| n/N (%) | 21/170 (12.4%) | 20/170 (11.8%) | 2/170 (1.2%) | 3/170 (1.8%) | |
For positive samples, viral load is expressed as the log10 number of copies of HEV-RNA per mL of fecal suspension if higher than the quantification limit of the real-time RT-PCR (>3.00log10 copies). Positive samples with viral load between 2.40 and 3.00log10 copies are expressed as the symbol +.
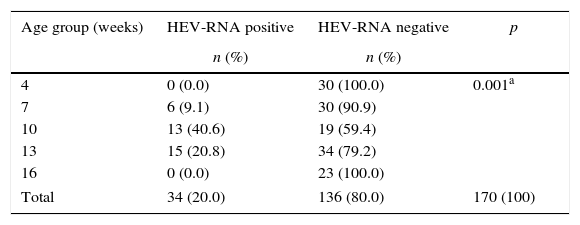
Table 2 shows the detection of HEV-RNA by age group. In the age group from 4 to 7 weeks, only 9.1% of pigs had detectable HEV-RNA, whereas 40.6% had detectable HEV-RNA at 10 weeks of age. These results show an approximate 7-fold higher risk of undergoing a HEV infection at the age of 10 weeks than at the ages of 4 or 7 weeks (OR=6.84, 95% CI 2.29–20.48).
HEV-RNA detection frequency in pigs from the rural area of Paraná State, South Brazil, by age group.
| Age group (weeks) | HEV-RNA positive | HEV-RNA negative | p |
|---|---|---|---|
| n (%) | n (%) | ||
| 4 | 0 (0.0) | 30 (100.0) | 0.001a |
| 7 | 6 (9.1) | 30 (90.9) | |
| 10 | 13 (40.6) | 19 (59.4) | |
| 13 | 15 (20.8) | 34 (79.2) | |
| 16 | 0 (0.0) | 23 (100.0) | |
| Total | 34 (20.0) | 136 (80.0) | 170 (100) |
Due to the low viral load of the majority of the positive samples and the difference between the detection limit of the screening real-time RT-PCR and the conventional nested RT-PCR, only 4 (11.8%; 4/34) samples could be amplified with conventional nested RT-PCR and subjected to direct sequencing. Among these samples, two were amplified with the set of primers resulting in a final product of 287nt, and three were amplified with the 384nt set. Sample 079 was amplified using both sets of primers on the conventional nested RT-PCR.
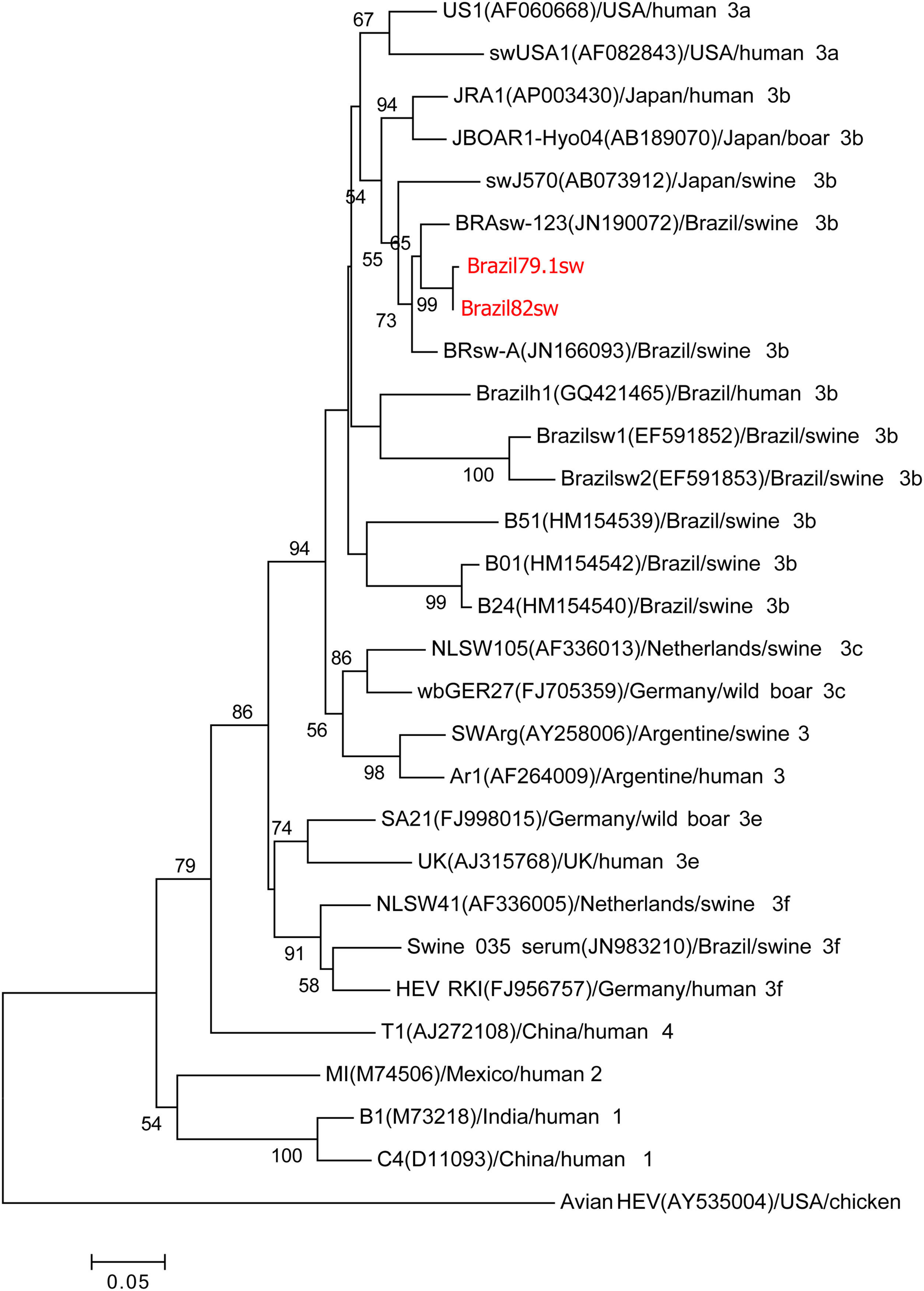
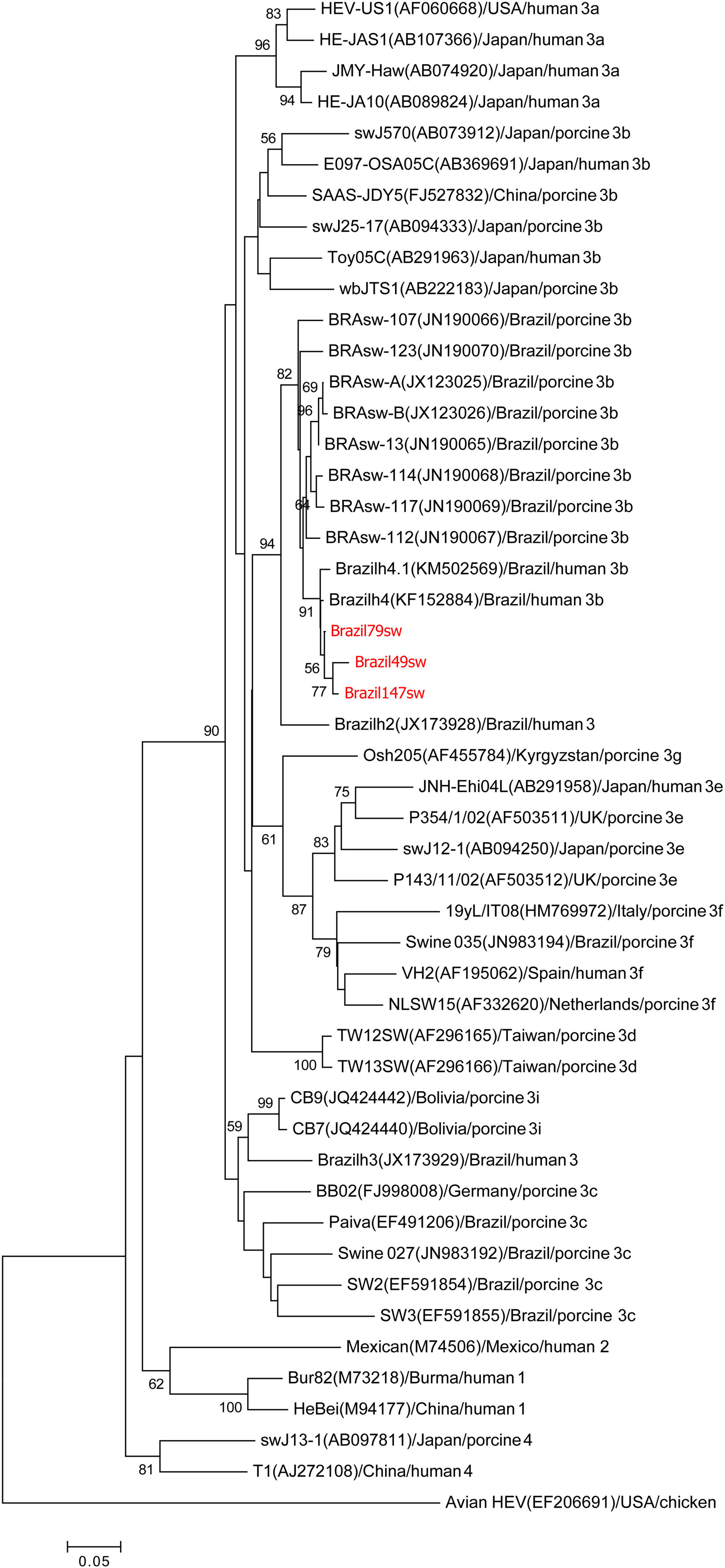
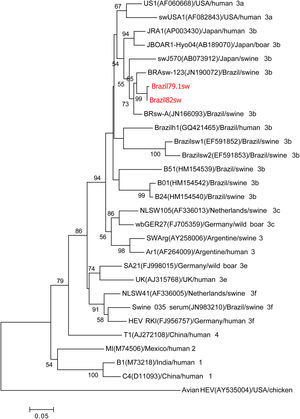
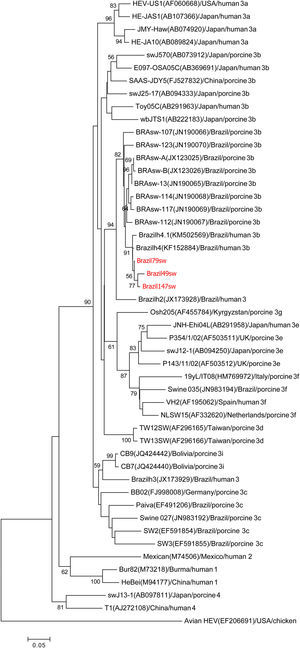
Phylogenetic analysisThe two samples from the 287nt ORF1 partial region shared 99% homology with each other and clustered with the genotype HEV-3b reference sequences in the phylogenetic analysis (Fig. 1). The three samples from the 384nt ORF2 partial region also grouped with the genotype HEV-3b reference sequences in the phylogenetic analysis. Nucleotide identity among these three samples ranged from 98% to 99% (Fig. 2).
Phylogenetic tree reconstructed by the neighbor-joining method with common 242-nt ORF1 sequences from 29 isolates, including 8 porcine isolates from Brazil, 1 human isolate from Brazil, and the 2 swine isolates described in this study, Brazil79.1sw and Brazil82sw (highlighted in red). The GenBank accession number in parentheses, the name of the country of origin, the species from which it was isolated, and the genotype/subtype of the isolate identify each viral strain. Bootstrap values of >50 are indicated for the major nodes as a percentage of the data obtained from 1000 replicates (bar, 0.02 substitutions per site). Major branches indicate genotypes. Avian HEV is the outgroup.
Phylogenetic tree reconstructed by the neighbor-joining method with common 304-nt ORF2 sequences from 49 isolates, including 13 porcine isolates from Brazil, 4 human isolates from Brazil, and the 3 swine isolates described in this study, Brazil79sw, Brazil49sw and Brazil147sw (highlighted in red). The GenBank accession number in parentheses, the name of the country of origin, the species from which it was isolated, and the genotype/subtype of the isolate identify each viral strain. Bootstrap values of >50 are indicated for the major nodes as a percentage of the data obtained from 1000 replicates (bar, 0.02 substitutions per site). Major branches indicate genotypes. Avian HEV is the outgroup.
The present data show a higher frequency of HEV infection (20.0%) in pigs than previously reported in Brazil. A study performed during 2009 in the same region with 170 fecal samples from 14 pig farms found HEV-RNA in 15.3% of samples.9 Another study that investigated serum, bile and fecal samples from 151 pigs from the eastern Brazilian Amazon, North Brazil, detected HEV-RNA in 9.9% of the animals.7 The higher infection rate observed in our study could be a result of the sanitary conditions of the small-scale pig farms in the rural area of Paraná State, where the contact of pigs of different ages may occur. However, it could also be due to the difference in methodology, since previous studies employed conventional RT-PCR techniques as opposed to the more sensitive real-time RT-PCR used in the present study. Additionally, the duplex RT-PCR technique employed in this study demonstrated the importance of using more than one set of primers and probes for higher detection during HEV screening, as only 20.6% of the positive samples had detectable HEV-RNA with both sets of primers and probes.
Previous studies reported that HEV infection in pigs was more frequent from 12 to 16 weeks of age and that at slaughter age (20–24 weeks), the animals had already developed anti-HEV antibodies.28 In the present study, HEV-RNA was more frequent in pigs aged 10 weeks (40.6%), although the frequency in pigs aged 13 was still high (20.8%). Nonetheless, none of the samples from the pigs aged 4 or 16 weeks tested positive. These observations confer with a study performed among swine herds in Rio de Janeiro, Southeast Brazil, showing that newborn pigs became susceptible to HEV between weeks 7 and 9, an age in which the serum levels of the maternal antibodies declined.5 These results are also in agreement with a study performed in Central Brazil in swine aged 20–30 weeks, with 81% of anti-HEV IgG positivity.29
The ingestion of raw or undercooked pork has been associated with HEV infection,30,31 and a probable zoonotic HEV transmission has been reported in Brazil.15 Additionally, there is a known risk of HEV transmission to people who come in contact with feces from infected pigs, which have been reported as an important source of infection for slaughterhouse workers and butchers and have been associated with infection in non-endemic regions.32,33 HEV has also been described in sewage samples from a slaughterhouse in Southeastern Brazil.6
The ORF1 HEV isolates found in this study shared 88% homology with a human HEV sequence from Brazil15 and 86% to 96% homology with swine HEV sequences from Brazil.5,8,9 The ORF2 HEV isolates shared 86–93% homology with human HEV sequences previously characterized by our research group in renal transplant recipients in Brazil16 and 83–97% homology with swine HEV sequences from Brazil.5,9 Among all compared HEV sequences, the highest homology (98–99%) was with human sequences recently isolated in Southeastern Brazil from a pediatric liver transplant recipient with chronic HEV infection.17
Although more studies are needed to elucidate the real impact of swine HEV infection and zoonotic transmission in Brazil, taken together, these results reinforce the hypothesis that domestic pigs may be an important source for human hepatitis E virus infection in this setting.
ConclusionsIn conclusion, this study confirms the circulation of the hepatitis E virus and shows a high frequency of HEV infection in pigs of different ages raised for slaughter in the rural area of Paraná State, South Brazil. The close similarity between the human HEV strains and those found herein indicates that adequate safety actions should be taken to prevent HEV infection when handling pigs and/or consuming pork.
Conflicts of interestThe authors declare no conflicts of interest.
This study was supported by the Fundação de Amparo à Pesquisa do Estado de São Paulo (Grant Nos. 2012/22925-3 and 2013.03701-0). We wish to thank Mr. Voitena, his family, and Veterinary Doctor Jessica Voitena for all their collaboration and technical assistance.