Malignant Catarrhal Fever (MCF) was investigated in the central nervous system of cattle with neurological syndrome. Two-hundred-ninety samples were analyzed by histology, and molecular methods to detect ovine herpesvirus type 2 (OvHV-2) were optimized and validated. The qualitative polymerase chain reaction (qualitative PCR) analytical sensitivity was 101DNA copies/μL and found 4.8% (14/290) positive for OvHV-2. The quantitative polymerase chain reaction (qPCR) analytical sensitivity was 100DNA copy/μL and 5.9% (17/290) positivity, with 47.1% (8/17) of the positive samples presenting histological evidence of non-purulent meningo-encephalitis. The qualitative PCR products (422bp of the ORF75 region) were sequenced and submitted to phylogenetic analysis. Identity matrices showed 100% similarity in OvHV-2 samples obtained in this study and those recovered from GenBank, corroborating other studies.
Encephalitis and encephalopathies have serious impact on public health, produce economic losses, and are a barrier to international trade.1 The differential diagnosis of bovine neurological syndrome is essential, and is challenging due to the difficulty in identifying potential agents and obtaining conclusive results. Diagnosis begins with investigations for the most probable agents, and, in the case of negative results, proceeding to include other pathogens.1
Malignant Catarrhal Fever (MCF) is a widespread disease that affects multiple systems and is often fatal to many artiodactyl species. It is caused by Herpesvirus of the family Gammaherpesvirinae and Macavirus, a group of antigenic and genetically related viruses, known as MCF viruses,2 which cause clinical or subclinical infection in susceptible animals.3–5 To date, ten MCF viruses have been identified, with the most frequent being ovine herpesvirus type 2 (OvHV-2), which causes MCF in sheep (SA-MCF).6
Diagnosis of SA-MCF is generally made from clinical history and signs, non-specific lesions observed during necropsy, histology, and epidemiologic data.7 Molecular methods are recommended, as they allow viral DNA detection both ante- and post-mortem, to identify the disease and assess its epidemiology.
Lack of awareness of MCF as a cause of neurological syndrome in cattle and the importance of the differential diagnosis of neurological diseases reinforces the need for optimization and validation of specific and sensitive diagnostic methods for the molecular identification of the OvHV-2.
This research aimed to evaluate suspected cases of MCF in cattle by clinical and anatomopathological evaluation and by molecular characterization in Brazil.
Materials and methodsSurvey designRetrospective molecular and histological analyses were made of 290 central nervous system (CNS) samples from cattle with neurological symptoms referred to the Centro de Pesquisa e Desenvolvimento de Sanidade Animal do Instituto Biológico for differential diagnosis of neurological syndrome. The samples came from several regions of Brazil, with 285 collected from January 2012 to March 2014, one in 2007, and four in 2009. All samples were negative for rabies virus, bovine herpesvirus types 1 and 5, bovine viral diarrhea, and Neospora caninum.
Standard virus selectionA sample of bovine CNS from Assis County, São Paulo State was used as virus standard. The sample was found MCF-positive on histological examination. The viral DNA was extracted using TRizol according to the manufacturer's instructions. Amplification of the ORF75 segment that encodes the phosphoribosylformylglycinamidine synthase (FGARAT) enzyme, participating in purine metabolism and production of viral tegument proteins, was conducted by PCR, using the primers MCF 556 and MCF 755, to amplify a 422 base pair (bp) segment. The positive PCR products were purified, sequenced, and identified as ovine herpesvirus-type 2 (OvHV-2). It was quantified using the QuantiFluor dsDNA System following the manufacturer's instructions. The purified DNA was used as standard to optimize and validate molecular methods.
To determine the sensitivity of the molecular methods, ten-fold serial dilutions of a positive sample (100–107DNA copies/μL) were performed in nuclease free water and in CNS negative for OvHV-2.
Extraction of nucleic acidThe DNA samples were extracted using TRizol, according to the manufacturer's instructions. The viral load was determined by comparing to the standard curve, expressed as DNA copies per gram tissue.
Optimizing and validating molecular methodsPrimers and an hydrolysis probe targeting the ORF75 regions of the OvHV-2 genome were used. For qualitative PCR, the primers used were MCF 556 (5′ GTC TGG GGT ATA TGA ATC CAG ATG GCT CTC 3′), MCF 755 (5′ AAG ATA AGC ACC AGT TAT GCA TCT GAT AAA 3′), and MCF 555 (5′ TTC TGG GGT AGT GGC GAG CGA AGG CTT C 3′).8 For qPCR, the primers used were forward oF-OvHV-2 (5′ TGG TAG GAG CAG GCT ACC GT 3′), reverse oR-OvHV-2 (5′ ATC ATG CTG ACC CCT TGC AG 3′), and the hydrolysis probe FAM/TAMRA oP-OvHV-2 (5′ TCC ACG CCG TCC GCA CTG TAA GA 3′).9
The temperature gradient was tested in the qualitative PCR with MCF 556 vs. MCF 755, MCF 556 vs. MCF 555, and MCF 755 vs. MCF 555 with various reagent concentrations and cycling conditions (data not shown).
Reaction conditions in both amplifications were adjusted to 12.5μL PCR Master Mix (Promega). The concentration of external and internal primers was 0.25μM (556 and 755 in the first amplified 422bp, 556 and 555 in the second 238bp). The volume of DNA in the first and second amplification was 2.5μL and 2μL, respectively, plus nuclease free water to a total volume of 25μL.
Quantitative PCR used SYBRGreen and TaqMan systems at primer concentrations of 200–1000nM. The hydrolysis probe was at 60–100nM concentration (data not shown). All reactions were carried out in a LightCycler 480 (Roche).
For the SYBRGreen assay, a commercial kit LightCycler 480 SYBR Green I Master (Roche Molecular Systems) was used with 10μL Master Mix, 2× concentration. The concentration of both primers was 500nM; 2.0μL DNA, and PCR-grade water to a total volume of 20μL.
The TaqMan system was implemented using the LightCycler 480 Probe Master (Roche Molecular Systems) with 10μL Master Mix, 2× concentration. The concentration of both primers was 500nM and the probe was 90nM; 2.0μL DNA and PCR-grade water to a total volume of 20μL.
Following the optimized amplification conditions (Table 1), the standard virus dilution in nuclease-free water, as well as in extracted DNA of CNS negative for OvHV-2, were submitted to three replications in three days, for validation and confirmation of results of the assay.10
Cycling and temperature conditions of the first and second amplification for qualitative PCR and both qPCR systems (SYBR Green and TaqMan Systems) for OvHV-2.
| Method | System | Primers | Initial denaturing | 30 cycles | Final extension | Curve of melting | ||
|---|---|---|---|---|---|---|---|---|
| DNA denaturing | Primer hybridization | Polymerization | ||||||
| Qualitative PCR | First amplification: MCF 556/755 | 95°C/3min | 95°C/30s | 60°C/30s | 72°C/30s | 72°C/3min | ||
| Second amplification: MCF 556/555 | 95°C/3min | 95°C/30s | 67°C/30s | 72°C/30s | 72°C/3min | |||
| Quantitative PCR | SYBR Green | oF-OvHV-2 | 95°C/5min | 40 cycles | 72°C 5s/65°C 1min/97°C continuous | |||
| 95°C/10s | 60°C/20s | 72°C/6s | ||||||
| TaqMan | oR-OvHV-2 | 95°C/5min | 95°C/10s | 60°C/20s | 72°C/6s | |||
A 422bp fragment from 13 positive samples was submitted to sequencing using the Applied Biosystems ABI 3500XL Genetic Analyser (Applied Biosystems). The quality of the sequences was assessed using Sequence Analyzer (Applied Biosystem) software. BioEdit v. 7.1.11,11 BLAST and ClustalW software were used to analyze the sequences. Alcelophine Herpesvirus-1 was used as outgroup. Molecular Evolutionary Genetics Analysis (MEGA) v. 6.012 was used for phylogenetic analysis. The tree was generated by a neighbor-joining algorithm using Kimura 2 evolutionary model parameters with 1000 bootstrap replicates.
HistologyThe samples were conventionally processed and stained with hematoxylin and eosin13 for histological examination.
ResultsStandard virusThe selected standard sample was the identified 798/07, collected in Assis County, São Paulo State, with MCF clinical and histological diagnosis (Fig. 3). The 422bp sequence presented 97–100% similarity to OvHV-2 from several regions throughout the world.
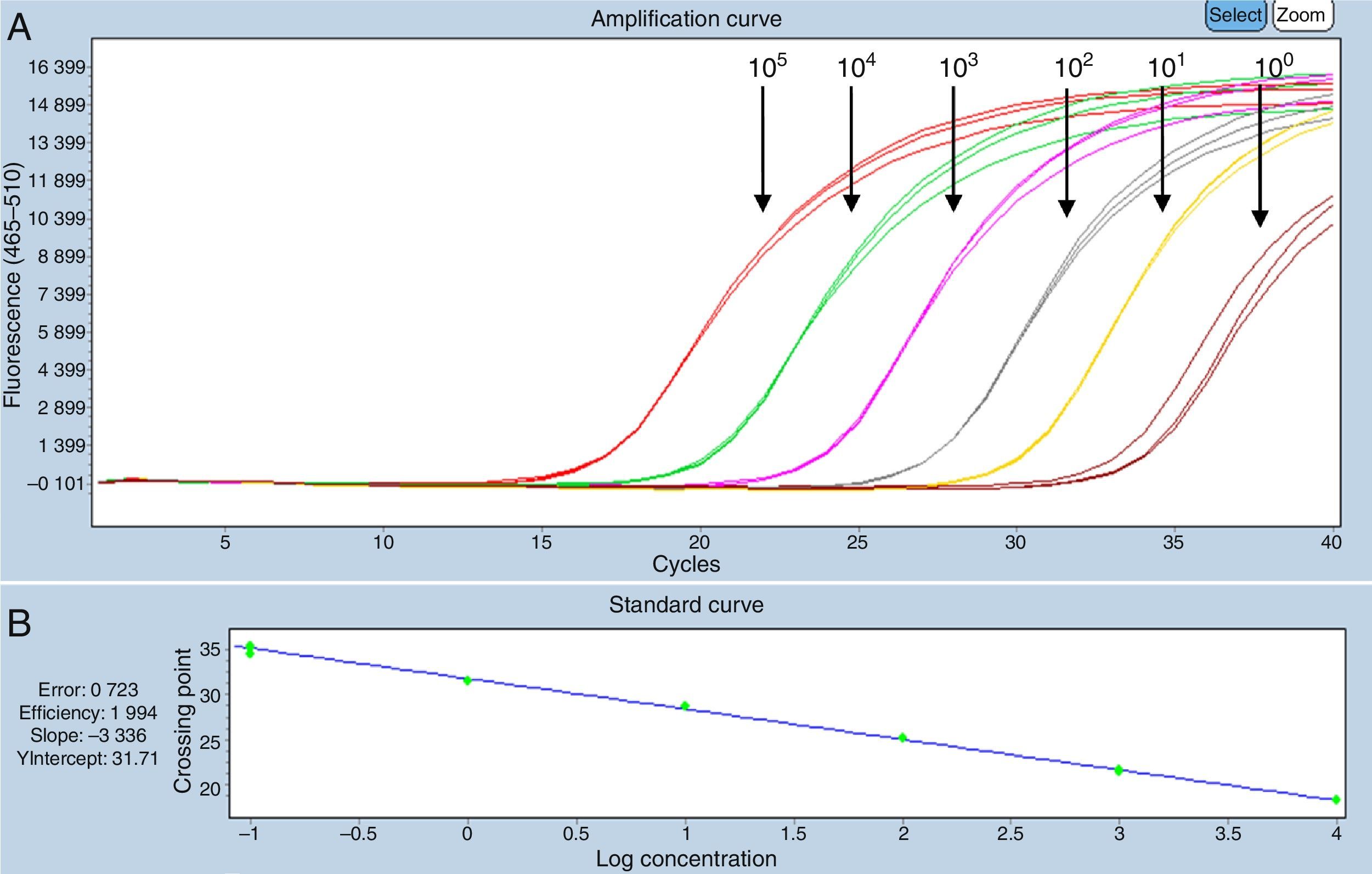
Optimizing and validating the molecular methodsThe qualitative PCR analytical sensitivity in the first amplification was 101DNA copies/μL and, in the second, was 102DNA copies/μL.
The detection threshold of both quantitative PCR systems was 100DNA copies/μL [quantification cycle (Cq) ∼35 cycles]. There were no differences between the results obtained from standard virus dilutions in extracts of DNA of CNS negative to OvHV-2 and in nuclease free water (Figs. 1 and 2).
Standard curve SYBRGreen system, dilutions in extracts of CNS of cattle negative to OvHV-2. (A) Serial dilutions 100–105DNA copies/μL. (B) Specificity of the reaction: efficacy=2.001, slope=−3.319. (C) Melting curve generated in the SYBRGreen system, dilutions in extracts of CNS of cattle negative to OvHV-2 with peak at Tm 86°C.
Of the bovine CNS samples collected from January 2012 to March 2014, 4.2% (12/285) were positive for OvHV-2 by qPCR. An additional four cases were confirmed in 2009 and one in 2007, totaling 5.9% (17/290) positivity. The qualitative PCR detected 4.8% (14/290) positive samples, and 2.8% (8/290) presented characteristic minor or greater grade histopathological changes (Fig. 3).
Histopathological alterations observed in MCF in tissues of animal 798/07. (A) Carotid rete mirabile with moderate mononuclear inflammatory infiltration and hyalinization of the tunica adventitia (H & E 200× magnification). (B) Cerebral cortex with moderate mononuclear perivascular cuffing in gray matter and mononuclear infiltrate in meninges (non-purulent meningo-encephalitis) (H & E 200× magnification). (C) Thalamus with intense mononuclear perivascular cuffing in the neuropil (H & E 100× magnification). (D) Kidney. Renal cortex with intense mononuclear inflammatory infiltrate and hyalinization in the tunica media and adventitia (H & E 100× magnification).
The agreement between qPCR (the most sensitive method) and qualitative PCR was 82.4% (14/17) and between qPCR and histology was 47.1% (8/17) (Table 2).
Sample identification for year collecting, age animals (in months), number of dead or sick animals in the herd, quantification cycle – Cq, DNA copies per tissue gram by qPCR, results of qualitative PCR, results of histology and country/state where the sample came from.
| Sample identification | Year | Age (months) | Number of dead or sick animals/herd | Quantification cycle (Cq) | DNA copies per tissue gram | Qualitative PCR | Histology | County/state |
|---|---|---|---|---|---|---|---|---|
| 798/07 | 2007 | 96 | 1/21 | 26.4 | 1.5×105 | POS | POS | Assis/SP |
| G355/09 | 2009 | 36 | 2/350 | 26.8 | 1.2×105 | POS | POS | NIa/PE |
| G86/09 | 2009 | 48 | 3/530 | 24.6 | 4.6×105 | POS | POS | NIa/PE |
| G251/09 | 2009 | 3 | NIa | 26.5 | 1.5×105 | POS | POS | Lagoa D’Ouro/PE |
| 1398/09 | 2009 | 56 | 1/150 | 30.9 | 3.9×104 | POS | NEG | Martinópolis/SP |
| 364/12 | 2012 | 38 | NIa | 29.7 | 1.7×104 | POS | POS | Socorro/SP |
| 11570/12 | 2012 | 84 | NIa | 35.2 | 0.9×103 | NEG | NEG | Caçapava/SP |
| 24221/12 | 2012 | 18 | 1/100 | 30.4 | 0.9×104 | POS | NEG | Ribeirão Preto/SP |
| 20428/13 | 2013 | 36 | NIa | 24.1 | 6.6×105 | POS | POS | Bujari/AC |
| 5587/13 | 2013 | 48 | NIa | 35.2 | 0.9×103 | NEG | NEG | Franca/SP |
| 6414/13 | 2013 | 24 | 1/29 | 32.5 | 2.9×104 | POS | NEG | Restinga/SP |
| 7983/13 | 2013 | 48 | 5/350 | 33.3 | 1.4×104 | POS | NEG | Itirapuã/SP |
| 8011/13 | 2013 | NIa | 12/3106 | 33.6 | 1.1×104 | POS | NEG | Queiroz/SP |
| 8012/13 | 2013 | 48 | 1/397 | 28.9 | 3.9×104 | POS | POS | Morungaba/SP |
| 8013/13 | 2013 | 5 | 1/254 | 33.5 | 1.0×104 | POS | NEG | Itaguaçu/ES |
| 25605/13 | 2013 | 48 | 1/350 | 33.7 | 0.8×104 | POS | POS | Silva Jardim/RJ |
| 2062/14 | 2014 | 36 | 3/144 | 35.0 | 1.0×103 | NEG | NEG | Itirapina/SP |
The viral load of samples associated with pathological evidence varied from 0.8×104 to 6.6×105 DNA copies/g tissue, and the viral load of samples without pathology varied from 0.9×103 to 3.9×104 DNA copies/g tissue (Table 2).
Samples of CNS, kidney, peripheral blood leukocytes, nasal swab, eyes, and lymph nodes of the animal identified as 20428/13 were collected. The tissues were stored separately until analysis, to prevent cross contamination, and analyzed separately to evaluate variations in positivity. All samples were positive with qualitative PCR and both qPCR systems. Pathological changes were found in the carotid rete mirabile and in kidney.
SequencingThirteen positive samples from cattle of different regions of Brazil were sequenced (Fig. 4). The identity among samples and between samples and sequences obtained from the GenBank database ranged from 90.9 to 100% (Fig. 4).
Location of the sequenced samples: A – 798/07; B – G251/09; C – 1398/09; D – 364/12; E – 24221/12; F – 20428/13; G – 6414/13; H – 7983/13; I – 8012/13; J – 8013/13; and K – 25605/13. Phylogram of the nucleotide sequences obtained in the current study aligned with OvHV-2 sequences available in GenBank. The tree was built by the neighbor-joining algorithm using the Kimura 2 evolutionary model with 1000 bootstrap replicates.
MCF is generally diagnosed by history, epidemiological data, clinical signs, and lesions observed during necropsy and microscopy. Occasionally, it is diagnosed by immunodiagnostics and detection of viral genomic DNA. Malignant Catarrhal Fever diagnosis in Brazil is usually made by clinical signs, necropsy, and histology.14–21 Such diagnoses are not always reliable, because MCF may be confused with other diseases, resulting in under-diagnosis, since there are no characteristic macro- or microscopic lesions, and some lesions can be observed only upon post-mortem examination. Molecular methods are reliable for MCF diagnosis in ante-mortem examinations (uncoagulated blood, ocular and nasal swabs), providing early detection of the virus to avoid economic loss, dissemination of disease within the herd, and mortality. Molecular methods allow post-mortem diagnosis in organs and fluids. Molecular methods are highly sensitive and specific and allow the diagnosis of MFC in the absence of clinical signs (latency), as may exist in chronic disease and in recovered animals.22–24
Both qualitative and quantitative PCR in this study showed higher sensitivity than reported in previous studies.9,24,25 The use of SYBRGreen for OvHV-2 diagnosis showed the feasibility of these molecular methods in routine laboratory diagnoses.
As expected, the number of positive samples was higher in both qPCR analyses (5.9%, 17/290) than in qualitative PCR (4.8% – 14/290), reflecting differences in analytical sensitivity. The samples with numbers of viral particles exceeding101DNA copies/μL presented typical MCF histopathologies (2.8% – 8/290) in CNS and kidney (Fig. 3). The qPCR detected twice the SA-MCF cases as found by histology, considered a gold standard method. These observations demonstrate the importance of implementing a more sensitive, specific, rapid, and safe molecular diagnosis method, allowing early diagnosis and easy OvHV-2 detection. This method allows detection of low viral loads in a range of organs and fluids.
This research also draws attention to the importance of correct collection and delivery of material for histological diagnosis. In most cases a pathologist does not receive the carotid rete mirabile tissue necessary for MCF diagnosis, making histologic confirmation difficult and leading to under-diagnosis. In this case, molecular diagnosis is fundamental, due to its ability to detect the agent in various organs and fluids. In addition, the use of both molecular tools and histology is useful to differentiate active from latent OvHV-2 infection, considering that inflammatory lesions with mononuclear infiltrate (Fig. 3) are related to the presence of a viral agent.
Most of the farms on which positive animals were found had cattle and sheep pastured together, which was a risk factor for SA-MCF.
The phylogenetic tree and the genetic identity among Brazilian samples and with those from other parts of the world showed no significant genomic differences, suggesting that the OvHV-2 diagnosed in Brazilian cattle was genetically similar to that found in other countries.
These epidemiological data are important for Brazilian Animal Health authorities planning strategies for prevention of MCF in cattle.
Conflicts of interestThe authors declare that there is no conflicts of interest.
Thanks to Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) of the Brazilian Ministry of Education and Culture for granting the Master's scholarship, and to the technicians of the Coordenadoria de Defesa Agropecuária do Estado de São Paulo (Secretary of Agriculture and Supply of São Paulo State) for providing the CNS samples.