The objective of this study was to characterize genotypically Malassezia spp. isolated from the external ear canal of healthy horses. Fifty-five horses, 39 (70.9%) males and 16 (29.1%) females, from different breeds and adults were studied. External ear canals were cleaned and a sterile cotton swab was introduced to collect cerumen. A total of 110 samples were cultured into Dixon medium and were incubated at 32°C for up to 15 days. Macro- and micromorphology and phenotypic identification were performed. DNA was extracted, strains were submitted to polymerase chain reaction technique, and the products obtained were submitted to Restriction Fragment Length Polymorphism using the restriction enzymes BstCI and HhaI. Strains were sent off to genetic sequencing of the regions 26S rDNA D1/D2 and ITS1-5.8S-ITS2 rDNA. Malassezia spp. were isolated from 33/55 (60%) animals and 52/110 (47%) ear canals. No growth on Sabouraud dextrose agar was observed, confirming the lipid dependence of all strains. Polymerase chain reaction-Restriction fragment length polymorphism permitted the molecular identification of Malassezia nana – 42/52 (81%) and Malassezia slooffiae – 10/52 (19%). Sequencing confirmed RFLP identification. It was surprising that M. nana represented over 80% of the strains and no Malassezia equina was isolated in this study, differing from what was expected.
Yeasts of the Malassezia genus are considered regular inhabitants of the cutaneous microbiome of animals and humans, but they may also be involved in several diseases, from cutaneous to systemic.1–5
In the last few years, molecular studies have increased the number of species classified in the genus Malassezia to 14: M. furfur, M. pachydermatis, M. sympodialis, M. globosa, M. obtusa, M. restricta, M. slooffiae, M. dermatis, M. japonica, M. nana, M. yamotoensis, M. caprae, M. equina and M. cuniculi.6,7
All species are lipophilic and lipodependent, with the exception of M. pachydermatis. In veterinary medicine, M. pachydermatis is the most commonly isolated species2,3,5,8; however, studies have shown that the lipodependent species may also constitute the microbiome and cause diseases in domestic and wild mammals.8–11
Surveys with horses and domestic ruminants showed that the occurrence of lipodependent species was significantly higher than that of M. pachydermatis, when it was reported the presence of M. furfur, M. slooffiae, M. obtusa, M. globosa, M. sympodialis and M. restricta.12
Phenotypical characterization is still accepted to diagnose clinical cases; however, lipodependent species may present similar biochemical and physiological behavior, leading to erroneous identifications. Therefore, studies have suggested the use molecular methods that allow proper differentiation among species.4,6,11
External otitis is an extremely serious disease in horses, with uni/or bilateral presentation and the potential to become a chronic condition. A significant portion of these cases are bacterial in origin, but some reports of mycotic otitis in equines caused by yeasts are available in the literature.8,13 However, published studies regarding the presence of Malassezia in the ear canal of horses are scarce.8,12
It is critical to carry out new surveys in this area that will permit better understanding of the distribution of Malassezia species in the microbiome of equine external ear canals.
Therefore, the aim of this study was to genotypically characterize Malassezia spp. isolated from the external ear canal of healthy horses.
Materials and methodsFifty-five adult horses from different breeds, 39 (71%) males and 16 (29%) females were studied. This study evaluated stalled horses from an equestrian society in the city of São Paulo, Brazil.
The external ear canals were cleaned with an alcohol–ether solution (1:1) and a sterile cotton swab was introduced to collect cerumen from both ears. A total of 110 samples were cultured in modified Dixon and Sabouraud dextrose agar, and the plates were incubated at 32°C for up to 15 days.6 The macro-and micromorphology of the isolates were observed by Gram stain. The phenotypic identification was based on the physiologic characteristics, such as the assimilation and growth in different lipid sources, the Tween 20, 40, 60, 80, and cremophor-EL. A catalase reaction, splitting of esculin and growth at 40°C were also tested.6
DNA was extracted by 10min ebullition. After that, the strains were submitted to polymerase chain reaction (PCR), using the following primers: forward, 5′-TAACAAGGATTCCCCTAGTA-3′ and reverse, 5′-ATTACGCCAGCATCCTAAG-3′.14
Amplification of the DNA target sequence was performed with the kit Platinum® Taq DNA Polymerase (Invitrogen™, Carlsbad, CA, USA), in final volume reactions of 50μL: 34.3μL DEPEC sterile water (dimetil pyrocarbonate); 5.0μL of 10× PCR buffer; 1.0μL of 10mM dNTP (deoxynucleoside triphosphate, 0.2mM final concentration of each deoxynucleoside); 1.5mM of MgCl2; 0.2μM of forward primer 26S-Fw; 0.2μM of reverse primer 26S-Rv; one unit of Platinum® Taq DNA Polymerase and 5.0μL of the extracted genomic DNA. The tubes were incubated in an Eppendorf Mastercycler Gradient® 5333 thermocycler (Eppendorf, Hamburg, Germany) at 94°C for 5min for initial denaturation of the genetic material and enzyme activation, followed by 30 amplification cycles: denaturation at 94°C for 45s, annealing at 55°C for 45s, extension at 72°C for 1min and a final extension at 72°C for 7min.14
The temperature was maintained at 4°C after PCR and the samples were stored at −20°C. After amplification, the PCR products were submitted to electrophoresis on agarose gel (1.5% diluted in TBE buffer), programmed at 100V for an hour. Following the electrophoresis, 0.5μg/mL ethidium bromide solution was used to stain the gel for 15min and the gel was visualized on an UV transilluminator and photographed using the Gel Logic 200 Kodak system™ (Eastman Kodak Co., Rochester, NY, USA). The size of the amplification was estimated according to a 100pb molecular weight marker (100pb DNA Ladder; Norgen, CA, USA). M. pachydermatis CBS 1696 (580bp) and Milli-Q were used as positive and negative controls, respectively.
The products obtained by PCR were submitted to restriction fragment length polymorphism (RFLP)4 using the restriction enzymes BstCI (Bio Labs®, Ipswich, MA, USA) and HhaI (Invitrogen™, Carlsbad, CA, USA).14
A volume of 21.5μL of PCR product was added to 3.5μL (10units) of restriction enzyme (0.5μL enzyme and 3.0μL buffer), up to a total volume of 25μL. Tubes were prepared with two different enzymes for each PCR sample, and incubated at 37°C for 3h.14 Products obtained by the RFLP were applied on 2.0% agarose gel diluted in TBE buffer. Electrophoresis was set at 100V for 1h and 30min. Markers of molecular weight and band visualization were performed according to the same method previously described.
Species confirmation was based on the random selection of four strains: two with M. nana- and two with M. slooffiae-like patterns obtained by RFLP, which were submitted for genetic sequencing. The PCR products were purified with the ExoSAP-IT system (GE Healthcare Bio-Sciences Ltd – USB Corporation, Cleveland, OH, USA), according to the manufacturer's instructions and quantified with Thermo Scientific NanoDrop™ 1000 Spectrophotometer (Thermo Fisher Scientific Inc, Wilmington, DE, USA).
For the sequencing, 30ng of the amplified product were added to 2μL of 5× sequencing buffer (Applied Biosystems; Carlsbad, CA, USA), 3.2pmol of primer, 2μL of ABI PRISM DyeTerminator Cycle Sequencing Ready Reaction kit (Big Dye v3.1, Applied Biosystems; Carlsbad, CA, USA) and a sufficient volume of DEPEC water was used to achieve a final volume of 10μL.
The primers ITS5 (5′-GGAAGTAAAAGTCGTAACAAGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) were employed for sequencing the region ITS1-5.8S-ITS2 rDNA; NL1 (5′-GCATATCAATAAGCGGAGGAAAAG-3′) and NL4 (5′-GGTCCGTGTTTCAAGACGG-3′) were employed for sequencing the region 26S rDNA D1/D2.
Sequencing was performed on an ABI-PRISM® 3130XL (Applied Biosystems, Carlsbad, CA, USA) automatic sequencer with the POP6 polymer. The nucleotide sequences were analyzed by the BLAST program15 (http://blast.ncbi.nlm.nih.gov/Blast.cgi?CMD=Web&PAGE_TYPE=BlastHome) to confirm product specificity and were then aligned with BioEdit Clustal X program on the 26S and ITS regions of the CBS-Knaw references strains16 (http://www.cbs.knaw.nl/): CBS7956 – M. slooffiae e CBS9557 – M. nana.
This project has been approved by Research Ethics Committee n° 024/10 CEP/ICS/UNIP.
ResultsMalassezia spp. were isolated from 33/55 (60%) animals and 52/110 (47%) ear canals. No growth on Sabouraud dextrose agar was observed, confirming the lipid dependence of all strains.
All strains were positive for the catalase reaction and capable of splitting esculin. Tween 20, 40, 60 and 80 were assimilated by 37/52 (71%) strains, but not cremophor-EL; some of these strains presented poor assimilation of the lipids, and the other 15/52 (29%) strains showed different patterns of assimilation, which did not permit phenotypic speciation.
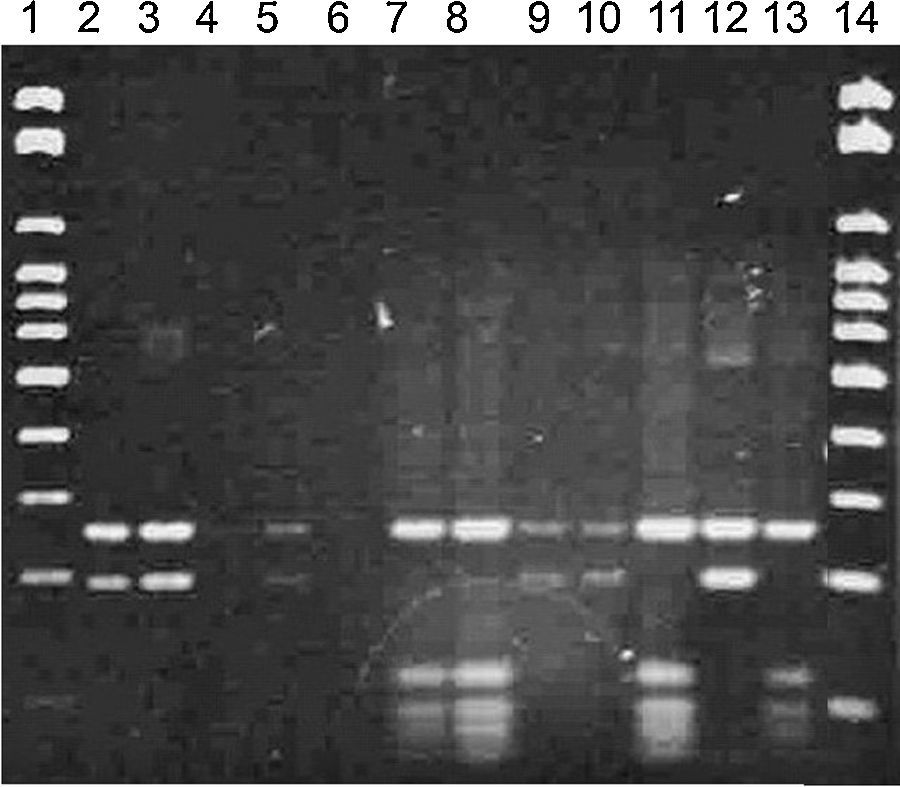
Bands between 500 and 600pb were obtained in the PCR technique, confirming all strains as Malassezia spp. (Fig. 1).
RFLP permitted the molecular identification of the 52 isolates from the ear canal in two species M. nana (42/52 – 81%) and M. slooffiae (10/52 – 19%) (Figs. 2 and 3).
Genetic sequencing of the regions 26S rDNA D1/D2 and ITS1-5.8S-ITS2 rDNA of the random four strains selected, when compared to CBS’ culture collection, confirmed RFLP identification, two of them were identified as M. nana and two as M. slooffiae.
DiscussionThe present study shows a high percentage of Malassezia spp. isolated from animals (60%) and ear canals (47%). The frequency of Malassezia spp. observed in the microbiome of the ear canals was similar, or even superior, to those mentioned by other studies that reported the presence of this yeast on the cutaneous surface of horses12,17,18 and other mammals.2,19,20 The high level of isolates in this study shows that lipodependent yeasts are part of the fungal microbiome in the ear canals of these horses.
Note that M. pachydermatis, which is considered to be the most common species from the cutaneous surfaces of animals,8,11 was not isolated in this study. In recent years, lipodependent species have been isolated from the skin and mucosa of healthy,10,19,20 as well as diseased animals.8,11,19–21
Although the incidence of external otitis in horses is low, the presence of Malassezia spp. in the microbiome of the ear canal suggests that these yeasts may cause infections when a host–parasite imbalance occurs, as has already been reported in equine cutaneous infections.22 The predisposing factors related to the transition of Malassezia spp. from a commensal organism to a pathogen are still poorly understood; being considered opportunistic fungi, these yeasts could over multiply and acquire a pathogenic characteristic.1 Mycotic otitis has already been indicated in horses,13 and the incidence of otitis in horses caused by Malassezia spp. could be underestimated because commercial veterinary laboratories do not use media with the addition of lipids for the determination of Malassezia spp., which might yield false-negative results in veterinary clinical samples.
Although phenotypic techniques are routinely used in clinical samples, in this study, these techniques failed to identify lipodependent Malassezia species. We observed that several strains did not present the expected patterns, which made their characterization, based on only phenotypic tests a lot more challenging.
In the RFLP technique, an enzyme cleaves the PCR product generating fragments of several sizes and consequently, bands with different molecular weights.1 In this research, the profiles obtained allowed the identification of the strains into two different species: M. nana and M. slooffiae that were identified by sequencing of the regions 26S rDNA D1/D2 and ITS1-5.8S-ITS2 rDNA.
In 2007, a new species named M. equina, was described and characterized by 26 rRNA gene sequencing.23 In the present study, genetic sequencing of the strains identified by PCR-RFLP confirmed the isolates as M. slooffiae and M. nana.
M. slooffiae has already been isolated from horses’ skin microbiome in a survey with 50 animals, as well as other lipodependent species: M. obtusa, M. globosa, M. sympodialis and M. restricta.12 Surprisingly, M. nana represented over 80% of the strains, and no M. equina was isolated in this study, differing from what was expected. It could be speculated that M. nana is a more prevalent species in the microbiome of the external ear canal of horses. However, to confirm such data, it is important to carry out new surveys in several different areas and increase the sampling; all of the animals evaluated in this study were placed in the same institution and subjected to similar husbandry practices.
M. nana has been isolated from cats with external otitis and bovines with or without otitis, suggesting that it may be part of the microbiome in some domestic animals; however, it has never been isolated from humans.24
FundingCapes-PROSUP fellowship, Federal Government, Brazil.
Conflicts of interestThe authors declare no conflicts of interest.