Solid-state fermentation can be used to produce feeds for ruminants, which can provide an enriched population of yeasts to improve ruminal fermentation. Fermentation of apple bagasse was performed to obtain a yeast-rich product, with the objective of isolating, identifying, and characterizing yeast strains and testing their capability to enhance in vitro ruminal fermentation of fibrous feeds. Yeasts were isolated from apple bagasse fermented under in vitro conditions, using rumen liquor obtained from cannulated cows and alfalfa as a fibrous substrate. A total of 16 new yeast strains were isolated and identified by biochemical and molecular methods. The strains were designated Levazot, followed by the isolate number. Their fermentative capacity was assessed using an in vitro gas production method. Strain Levazot 15 (Candida norvegensis) showed the greatest increase in gas production (p<0.05) compared with the yeast-free control and positively affected in vitro ruminal fermentation parameters of alfalfa and oat straw. Based on these results, it was concluded that the Levazot 15 yeast strain could be potentially used as an additive for ruminants consuming high-fiber diets. However, further studies of effects of these additives on rumen digestion, metabolism, and productive performance of ruminants are required.
A search for alternative, low-cost feed sources that can replace higher-cost conventional feeds is one of the main goals of animal nutritionists.1,2 The solid-state fermentation (SSF) process involves fermentation of solid substrates in the absence (or near absence) of free water and is frequently used for substrates containing enough moisture to support the development of fermenting microorganisms.3 Because SSF stimulates the growth of microorganisms, this process offers numerous opportunities for processing of agro-industrial residues, such as protein enrichment of sugarcane.4 Apple bagasse is a residue produced during extraction of apple juice. As an agricultural byproduct, apple bagasse is rich in soluble carbohydrates and pectins and thus can be utilized, after an SSF process, as a feed for ruminants. The fermentative process enhances the nutritive value of apple bagasse by promoting the growth of microorganisms, which increase its protein content and improve its digestibility.5 The use of yeasts such as Saccharomyces cerevisiae as feed additives has been shown to improve the health and productivity of ruminants6,7 and to offer a natural way to manipulate animal productivity by favorably modifying microbial fermentation and improving dry matter and neutral detergent fiber (NDF) digestibility,8 feed consumption,9 milk production, and live weight gain.10 A previous in vitro study has demonstrated that the addition of fermented apple bagasse (FAB) to ruminal fermentation of alfalfa increased the viable yeast counts and concentration of lactic acid in the ruminal medium within 24h.11 Other studies have shown that when certain yeast strains are provided, rumen conditions are improved.12,13 Similarly, significant increases were found in the in vitro forage degradability when rice bran was treated with Candida utilis, and the author attributed this effect to the stimulation of rumen microbes by the yeast.14 Marrero et al.15 demonstrated that supplemented yeasts were able to survive for 24h under the rumen conditions. These results confirm that certain microorganisms, when placed in a new habitat, are able to exploit the resources of the environment in which they are inoculated, as, for example, yeasts utilize the scarce oxygen present in the rumen, thus favoring anaerobic conditions.16 Based on the assumption that the addition of FAB would improve the ruminal fermentation, specifically due to the yeasts present in FAB, the objective of this study was to isolate, identify, and characterize yeast strains from FAB for their potential use as microbial additives in ruminant production systems.
Materials and methodsFermented apple bagasseApple bagasse was obtained from Confrutta, S.A. (Chihuahua, Mexico) and fermented as described by Castillo-Castillo et al.5 Briefly, apple bagasse was ground and mixed with urea, ammonium sulfate, and a mineral salt mixture containing macro- and microelements, which were added to final concentrations of 1.5%, 0.2%, and 0.5%, respectively.17 Then, 342g of the sample was placed into a sterile 500-mL Erlenmeyer flask for SSF. The flasks were plugged with cotton and incubated under static conditions at 32°C for 48h.
In vitro ruminal fermentation of apple bagasseNon-lactating, rumen-cannulated Holstein dairy cows (average body weight: 550±25.5kg, n=3) were used as donors of ruminal liquor. The cows were fed twice daily with 4.0kg of a concentrate (51.0% corn, 23.5% wheat bran, 10% cottonseed meal, 8.49% corn gluten meal, 2.0% sugarcane molasses, 1.5% soybean meal, 1.0% bypass fat, 0.8% CaCO3, 0.5% urea, 0.5% animal fat, 0.2% NaCl, and 0.5% trace mineral and vitamin premix) and 4kg of corn silage (on a dry matter basis). The rumen fluid was sampled at 6:00 a.m., before the morning feeding, and transferred to the laboratory in hermetically sealed sterile bottles. The ruminal liquor was filtered through six layers of cheesecloth under complete CO2 atmosphere to provide anaerobic conditions. Fermentation was performed in 20 serum flasks (250mL) at 39°C with mechanical agitation. The filtered rumen fluid was mixed with a buffer solution at a ratio of 1:2 (50:100mL) and added to a mixed substrate of FAB and alfalfa hay (50:50 ratio), which were previously milled and sieved through a 2-mm sieve before their transfer to the flasks for fermentation. After 24h, the flasks were withdrawn from the incubator, and their entire contents were collected, homogenized, and filtered through six layers of cheesecloth. The filtrates were used for inoculation of a medium in roll tubes, as described below. The chemical composition of the alfalfa (% of dry basis) was as follows: organic matter (OM), 89.07; ash, 10.93; crude protein (CP), 18.02; NDF, 65.6; acid detergent fiber (ADF), 39.0; and ether extract (EE), 2.02. The chemical composition of the fermented apple bagasse (% of dry basis) was as follows: OM, 89.94; ash 10.05; CP, 35.05; NDF, 48.31; ADF, 37.54; and EE, 5.2.
Culture of microorganismsYeasts were cultivated under strict anaerobic conditions according to the method described by Hungate.18 Roll tubes were inoculated in triplicate with three dilutions (104, 105, and 106) of the filtered contents of the ruminal fermentation flasks, according to Caldwell and Bryant,19 and incubated at 39°C for 24h. The isolation medium included malt extract agar (Difco™, Sparks, MD, USA) supplemented with 0.01g/L of chloramphenicol. The yeast isolates present in the ruminal ecosystem were obtained from colonies that grew in the roll tubes with the highest dilution, following the method of Marrero et al.20 Cultures with different macroscopic characteristics were inoculated and incubated following the method described by Marrero et al.21
Biochemical and microscopic characterization of Levazot strainsTo ensure that the isolated yeasts do not belong to the genus Saccharomyces, the isolates (designated Levazot, followed by the isolate number) were grown at 30°C for 72h on the following specific medium for non-Saccharomyces yeasts: malt agar extract (3.2g/L), peptone (1.8g/L), dextrose (10g/L), K2HPO4 (1g/L), NH4Cl (0.5g/L), CuSO4 (5%, w/v), and bacteriological-grade agar (19g/L).21S. cerevisiae strain L/25-7-13 obtained from the Cuban Institute for Research on Sugarcane Derivatives, National Center of Animal Health, was used as a control. To determine their oxygen requirements, the strains were grown in malt extract broth, on malt extract agar, and in thioglycollate medium at 30°C for 48h. Urea hydrolysis, starch fermentation, and high osmotic pressure tests were performed according to the methods described by Marrero et al.20 To observe the formation of pseudomycelium, mycelium, and ballistospores, the Levazot strains were seeded in Petri dishes with malt extract agar and incubated at 25°C for 5 days.
Molecular characterizationGenomic DNA was isolated using the PureLink™ Genomic DNA Mini Kit (Invitrogen, Carlsbad, CA, USA), according to the manufacturer's instructions. The 18S rRNA gene was amplified using the oligonucleotides nu-SSU-0817-5′ (5′-TTAGCATGGAATAATRRAATAGGA-3′) and nu-SSU-1536-3′ (5′-ATTGCAATGCYCTATCCCCA-3′), according to Borneman and Hartin.22 Each polymerase chain reaction (PCR) contained 5μL of 10× High Fidelity PCR buffer, 2μL of 50mM MgSO4, 2μL of 10mM deoxynucleotide triphosphates, 10pmol each primer NL1 and NL4, 200ng of DNA, and 1 U of Platinum® Taq DNA Polymerase, High Fidelity (Invitrogen) in a final volume of 25μL. Amplification was carried out as follows: initial denaturation at 95°C for 5min, followed by 35 cycles of 15s at 94°C, 25s at 54°C, and 20s at 68°C, and a final extension at 68°C for 10min. The PCR products were purified and sequenced using an ABI PRISM® 3100 Genetic Analyzer (Perkin Elmer). The sequences were submitted to the GenBank database using the BankIT program (National Center for Biotechnology Information, USA) and analyzed using the BLASTn algorithm.23 Phylogenetic analysis was performed using the PHYLIP24 and ClustalW programs.25 In addition to the yeasts isolated in this study, 18S rRNA gene sequences from 15 closely related yeasts were included in the analysis. The yeasts were Candida norvegensis (AF201302.1), C. albicans (KC936147.1), C. tropicalis (JQ008834.1), C. xestobii (AB013517.1), C. rugopelliculosa (EF550376.1), C. carpophila (AJ508270.1), C. fermentati (AY553853.1), C. dubliniensis (X99399.1), C. viswanathii (EU589205.1), Issatchenkia orientalis (EF550360.1), I. occidentalis (AB053240.2), I. scutulata (EF550381.1), I. terricola (EF550371.1), Pichia membranaefaciens (X96453.1), and P. nakasei (EF550386.1). A phylogenetic tree was generated using Kimura's two-parameter correction model for genetic distances26 and the neighbor-joining method.27
In vitro gas productionThe fermentative capacity of the isolated yeasts was assessed using the in vitro gas production method of Theodorou et al.28 The isolates were grown in 10mL of malt extract broth at 30°C for 24h on a shaker and then subcultured (1:40 dilution) in the same medium at 30°C for 24h. The gas production test was performed in four replications for each yeast strain as follows: 6mL of a culture (4.5×107cells/mL) was mixed with 24mL of the fermentation medium, and the mixture was transferred to a 50-mL glass bottle containing 0.3g of alfalfa hay and 0.3g of oat straw as substrates. The chemical composition of the oat straw (% of dry basis) was as follows: OM, 88.7; CP, 5.3; ADF, 54.5; and NDF, 69.1. The bottles were incubated at 39°C with shaking at 120rpm. The gas levels were measured after 48h of fermentation by bubble displacement in a 10-mL syringe.
Statistical analysisIn vitro gas production data were analyzed by a fully randomized analysis of variance using SAS PROC MIXED29 with a model that included the Levazot strains as fixed effects, the S. cerevisiae strain (control), a blank without yeasts, the fermentation time, and the interaction of both (strains and time). The random effect was the bottle (experimental unit) nested into the yeast strain. Comparison of means was performed using the PDIFF procedures,29 and the differences were determined to be statistically significant at p<0.05 or as indicated.
Results and discussionIsolation and characterization of Levazot strainsSixteen yeast strains resistant to the ruminal ecosystem were isolated using in vitro ruminal fermentation of FAB combined with alfalfa hay. The strains were designated Levazot, followed by the isolate number (Fig. 1). Based on the colony characteristics, the Levazot strains were divided into two groups. Group I contained two strains, Levazot 9 and Levazot 25, which developed white-colored, flat colonies, with irregular edges and a central papilla surrounded by a halo (Fig. 1). Group II contained Levazot strains 2, 3, 7, 8, 11, 13, 14, 15, 16, 19, 23, 29, 30, and 31, which formed white, creamy, flat colonies, with irregular edges, a papilla surrounded by a creamier circle, and a small downiness on the edges (Fig. 1). Based on the microscopic observations, only Levazot strains 2, 3, and 4 developed true mycelia, while the rest formed pseudomycelia, and none formed ballistospores. The biochemical test in which the oxygen relation was measured showed that in liquid medium all Levazot strains grew on the surface, forming a thin ring layer. The growth was from the surface to the inside of the medium, and all strains formed sediment (data not shown). The biochemical tests showed that all Levazot strains were negative for urea hydrolysis and positive for starch fermentation. Also, all the strains were able to grow in the specific culture medium for non-Saccharomyces yeasts and under high osmotic pressure conditions.
Molecular characterization of Levazot yeast strainsTo determine the taxonomic identity of the Levazot yeasts, the 18S rRNA gene was amplified from each isolate and sequenced, and the sequences were submitted to the GenBank database (Table 1). Bioinformatic analysis using BLASTn showed that all Levazot yeast isolates had 99–100% sequence identity with C. norvegensis and I. orientalis. To determine the genetic distances between the Levazot strains, C. norvegensis, and I. orientalis, a phylogenetic analysis was performed (Fig. 2), which assigned all Levazot strains into clade A (100% bootstrap values), i.e., the same clade where C. norvegensis and I. orientalis belonged. Clade A was divided into groups A.I, which included only Levazot 29, and A.II (42.9% bootstrap value), in which the rest of the Levazot strains were clustered. In the latter group, only Levazot 31 was placed in a separated subclade (A.II.1). Subclade A.II.2, with a 37.6% bootstrap value, was separated into two subgroups, A.II.2.1, with only Levazot 19, and A.II.2.2, which was further divided into two subgroups, A.II.2.2.1 and A.II.2.2.2. In the A.II.2.2.1 subgroup (20.4% bootstrap value), Levazot strains 2, 3, 7, 8, 9, 11, 15, 16, 23, and 25 were clustered with C. norvegensis, while Levazot 13, Levazot 14, and I. orientalis were grouped into A.II.2.2.2, with a 14.2% bootstrap value. According to the Candida classification by Cardenas,30C. norvegensis has the ability to assimilate starch, which agrees with the results presented in Table 1, but is not able to hydrolyze urea. None of the strains isolated in this study was ureolytic, but all fermented starch, unlike I. orientalis, which does not assimilate starch and is variable for hydrolysis of urea.30 Taken together, it is possible to infer, based on the facts that the genetic distances between Levazot strains 2, 3, 7,9, 15, 16, and 23 and C. norvegensis are very small and the strains have the same biochemical characteristics, that these new yeast strains belong to C. norvegensis, a species commonly isolated from cactus rots31–33 and occasionally isolated from human clinical samples.34
Identity of Levazot strains with yeast reported into GenBank, grouped by phylogenetic clade tree.
| Strain (GenBank Accession No.) | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Phylogenetic Clade | A. II.2.2.1 | A.I | A.II.I | A.II.2.1 | A.II.2.2.2 | |||||||||||
| Yeast (GenBank Accession No.) | Levazot 30 (JQ519373.1) | Levazot 11 (JQ519364.1) | Levazot 8 (JQ519362.1) | Levazot 25 (JQ519371.1) | Levazot 2 (JQ519358.1) | Levazot 23 (JQ519371.1) | Levazot 16 (JQ519368.1) | Levazot 7 (JQ519361.1) | Levazot 15 (JQ519367.1) | Levazot 9 (JQ519363.1) | Levazot 3 (Q519359.1) | Levazot 29 (JQ519372.1) | Levazot 31 (JQ519374.1) | Levazot 19 (Q519369.1) | Levazot 14 (JQ519366.1) | Levazot 13 (JQ519365.1) |
| C. norvegensis AF201302.1 | 100 | 100 | 100 | 99 | 100 | 100 | 99 | 100 | 99 | 100 | 99 | 100 | 100 | 100 | 99 | 99 |
| I. orientalis EF550360.1 | 100 | 100 | 100 | 99 | 100 | 100 | 99 | 100 | 99 | 100 | 99 | 100 | 100 | 100 | 99 | 99 |
| C. albicans KC936147.1 | 92 | 92 | 92 | 92 | 91 | 91 | 92 | 97 | 92 | 91 | 92 | 92 | 92 | 92 | 91 | 92 |
| I. occidentalis AB053240.2 | 98 | 98 | 98 | 98 | 98 | 98 | 98 | 98 | 98 | 98 | 98 | 98 | 98 | 98 | 98 | 98 |
| I. scutulata EF550381.1 | 98 | 98 | 98 | 98 | 98 | 98 | 98 | 97 | 97 | 98 | 98 | 98 | 98 | 98 | 97 | 97 |
| I. terricola EF550371.1 | 97 | 97 | 97 | 97 | 96 | 96 | 97 | 92 | 96 | 96 | 97 | 92 | 97 | 97 | 97 | 96 |
| C. tropicalis JQ008834.1 | 92 | 92 | 92 | 92 | 91 | 91 | 92 | 92 | 92 | 91 | 92 | 93 | 92 | 92 | 91 | 92 |
| C. xestobii AB013517.1 | 93 | 92 | 92 | 92 | 92 | 92 | 93 | 97 | 92 | 92 | 93 | 98 | 93 | 93 | 92 | 93 |
| P. membranaefaciens X96453.1 | 98 | 98 | 98 | 98 | 98 | 98 | 98 | 98 | 98 | 98 | 98 | 98 | 98 | 98 | 98 | 98 |
| C. rugopelliculosa EF550376.1 | 98 | 98 | 98 | 98 | 98 | 98 | 98 | 98 | 97 | 98 | 98 | 98 | 98 | 98 | 97 | 97 |
| C. carpophila AJ508270.1 | 92 | 92 | 92 | 99 | 91 | 91 | 92 | 91 | 91 | 91 | 92 | 92 | 92 | 92 | 91 | 92 |
| C. fermentati AY553853.1 | 92 | 92 | 92 | 92 | 92 | 92 | 92 | 92 | 92 | 92 | 92 | 93 | 92 | 92 | 92 | 92 |
| C. dubliniensis X99399.1 | 92 | 92 | 92 | 92 | 91 | 91 | 91 | 92 | 91 | 91 | 92 | 92 | 92 | 92 | 90 | 92 |
| C. viswanathii EU589205.1 | 92 | 92 | 92 | 92 | 91 | 91 | 92 | 92 | 92 | 91 | 92 | 92 | 92 | 92 | 91 | 92 |
| P. nakasei EF550386.1 | 98 | 98 | 98 | 98 | 98 | 98 | 98 | 98 | 97 | 98 | 97 | 98 | 98 | 98 | 97 | 97 |
| S. cerevisiae JQ409454.1 | 93 | 93 | 93 | 93 | 92 | 92 | 93 | 92 | 92 | 92 | 93 | 93 | 93 | 93 | 92 | 92 |
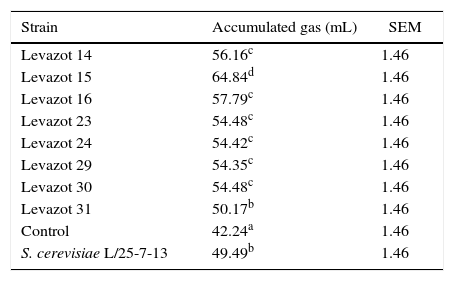
To select the Levazot strain with the best fermentative capacity, the isolates were individually incubated with alfalfa hay and oat straw as substrates. The gas levels were measured at 48h of fermentation (Table 2). All Levazot strains and S. cerevisiae had higher gas production levels than that observed in the no-yeast control (p<0.05). This implied that these yeasts stimulated the process of fermentation. At the same time, Levazot strains 13, 14, 15, 16, 23, 29, 30, and 31 showed higher gas production levels than that of S. cerevisiae (p<0.05). Because these Levazot strains showed similar behaviors when using alfalfa hay as a substrate, the same procedure was performed with oat straw, which is a lower-quality substrate than alfalfa. The results are shown in Table 3. Levazot 31 was the only strain that showed no statistical difference with S. cerevisiae; the rest had higher gas production levels (p<0.05). These results are similar to the data reported by Marrero et al.20 for Levica yeast strains isolated from the rumen, which produced more gas than S. cerevisiae with Cynodon nlemfuensis as a substrate. The statistical analysis confirmed that Levazot 15 demonstrated the highest gas production levels (p<0.05) with both alfalfa hay and oat straw. Due to this, Levazot 15 is considered the most promising yeast strain to be used as a microbial additive to improve digestive fermentation of ruminants fed fibrous diets. The higher gas production level obtained with this strain is probably due to an increase in propionic acid synthesis,35 which facilitates the formation of carbon dioxide (CO2) via the succinate–propionate metabolic pathway.36In vivo and in vitro studies have shown increased rates of cellulose and NDF degradation in the presence of yeasts,37,38 which could explain the increase in the gas production level found in this work. Similar results were reported by Tang et al.,39 who used straw of several cereals as substrates.
Fermentative capacity of Levazot strains by gas production at 48h of fermentation using alfalfa hay as substrate.
| Strain | Accumulated gas (mL) | SEM |
|---|---|---|
| Levazot 2 | 78.88bc | 2.55 |
| Levazot 3 | 81.71bcd | 2.55 |
| Levazot 7 | 77.22bc | 2.55 |
| Levazot 8 | 81.38bcd | 2.55 |
| Levazot 9 | 80.96bcd | 2.55 |
| Levazot 11 | 77.55bc | 2.55 |
| Levazot 13 | 83.79cdef | 2.55 |
| Levazot 14 | 89.78fg | 2.55 |
| Levazot 15 | 97.6h | 2.55 |
| Levazot 16 | 89.87fg | 2.55 |
| Levazot 23 | 90.78fg | 2.55 |
| Levazot 29 | 88.53efg | 2.55 |
| Levazot 30 | 92.36gh | 2.55 |
| Levazot 31 | 91.53gh | 2.55 |
| Control | 53.40a | 2.55 |
| S. cerevisiae L/25-7-13 | 75.14b | 2.55 |
Means with different superscript letters are significantly different (p<0.05).
SEM, standard error of means.
Fermentative capacity of Levazot strains by gas production at 48h of fermentation using oat straw as substrate.
| Strain | Accumulated gas (mL) | SEM |
|---|---|---|
| Levazot 14 | 56.16c | 1.46 |
| Levazot 15 | 64.84d | 1.46 |
| Levazot 16 | 57.79c | 1.46 |
| Levazot 23 | 54.48c | 1.46 |
| Levazot 24 | 54.42c | 1.46 |
| Levazot 29 | 54.35c | 1.46 |
| Levazot 30 | 54.48c | 1.46 |
| Levazot 31 | 50.17b | 1.46 |
| Control | 42.24a | 1.46 |
| S. cerevisiae L/25-7-13 | 49.49b | 1.46 |
Means with different superscript letters are significantly different (p<0.05).
SEM, standard error of means.
Thus, 16 new yeast strains (designated Levazot) were isolated and identified using in vitro ruminal fermentation of fermented apple bagasse in combination with alfalfa hay. Of these, strain Levazot 15 (C. norvegensis) showed the highest level of gas production in the in vitro tests with both alfalfa hay and oat straw, suggesting that this yeast strain enhanced ruminal digestion to a larger extent than the other strains. Therefore, we conclude that Levazot 15 could potentially be used as an additive to ruminant diets containing a high proportion of forage. Further studies are required on rumen digestion and metabolism and on productive performance of ruminants fed high-forage diets supplemented with these isolated yeast strains.
Conflict of interestThe authors declare no conflicts of interest.