The distinction between healthcare-associated MRSA (HA-MRSA) and community-associated MRSA (CA-MRSA) infections has become increasingly blurred. We assessed the molecular characterization and antimicrobial resistance profile for MRSA isolates from blood. Most of all (81.9%) isolates are related to known HA-MRSA and CA-MRSA epidemic lineages, such as, USA300, USA400, USA600, USA800 and USA1100. This is the first multicenter study in Rio de Janeiro.
Over the years, the distinction between healthcare-associated MRSA (HA-MRSA) and community-associated MRSA (CA-MRSA) has become increasingly blurred, with a growing number of reports indicating that CA-MRSA strains are spread in hospital settings and replacing traditional HA-MRSA strains, including in Brazil.1,2 HA-MRSA strains typically have a multidrug-resistant phenotype, whereas CA-MRSA strains are often susceptible to a variety of non-β-lactam antimicrobials.1 On the other hand, this resistance pattern is changing in the last decade. Thus, some HA-MRSA isolates are susceptible to many non-β-lactam antibiotics, including the isolates related USA800 clone, for example.1,3 The in vitro antimicrobial susceptibility to clindamycin and/or sulfamethoxazole/trimethoprim is a relevant characteristic described in strains with SCCmec IV.2 The genotyping of Brazilian MRSA isolates allowed a better understanding of the dissemination patterns of the epidemic clones, such as, typically HA-MRSA and CA-MRSA.4 The known virulence factor the exoprotein Panton-Valentine leucocidin (PVL) is traditionally seen in CA-MRSA,5 however it has already been identified in methicillin-susceptible S. aureus and some rare HA-MRSA isolates.1,6 The increasing number of reported infections caused by CA-MRSA strains in Brazil justifies the use of surveillance studies to help in understanding how these strains arrive and circulate in healthcare settings.2 Furthermore, recently, Rossi and colleagues7 reported a case of a Brazilian patient with a bloodstream infection caused by USA 300-related isolate, designated BR-VRSA, that acquired the vanA gene cluster during antibiotic therapy becoming resistant to vancomycin.
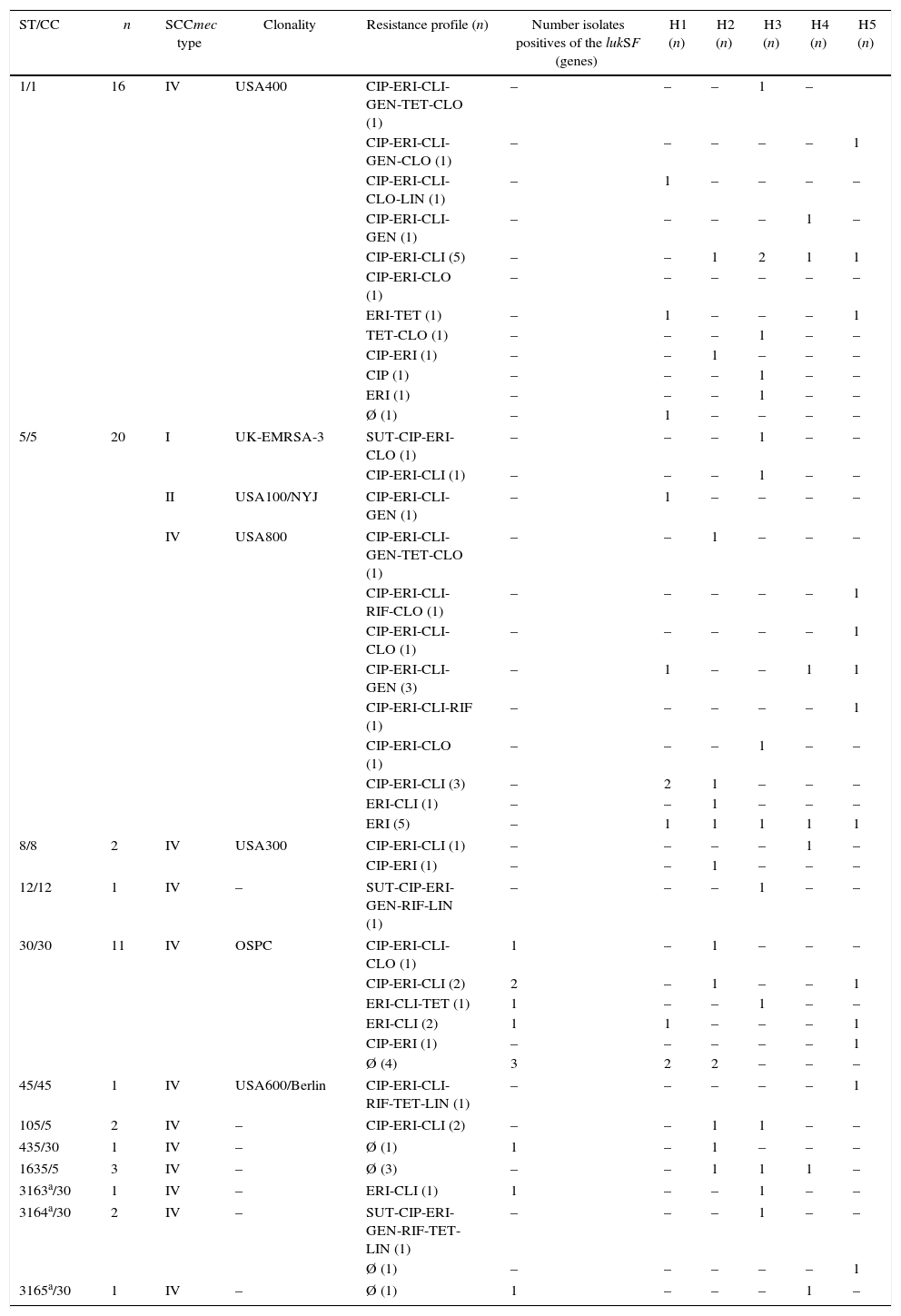
The present study analyzed the genetic background and resistance profile of MRSA isolates recovered from blood between March 2011 and May 2012. The inclusion criterium for the MRSA isolates studied here was the presence of susceptibility to the non-β-lactam antibiotics trimethoprim-sulfamethoxazole and/or clindamycin. The 61 MRSA blood isolates were collected from patients in five hospitals (H) in Rio de Janeiro, Brazil: H1 (n=11; 18.1%), H2 (n=14; 22.9%), H3 (n=16; 26.2%), H4 (n=7; 11.5%) and H5 (n=13; 21.3%), the last one is a pediatric unit. Using disk diffusion test,8 all MRSA isolates selected displayed high percentage of susceptibility to non-β-lactams.2,8 The following resistance rates was observed: 59.0% for clindamycin (CLI), 78.7% for erythromycin (ERI) and 60.6% for ciprofloxacin (CIP). Lower resistance rates were observed for chloramphenicol (CLO), 19.7%; gentamicin (GEN), 16.4%; rifampicin (RIF), 8.2%; tetracycline (TET), 11.5%; trimethoprim-sulfamethoxazole (SUT) and linezolid (LIN), 4.9%. Resistance to three or more non-β-lactam antibiotics was observed in 55.7% of the total isolates and only in H3 was observed strains resistant to SUT. None of the isolates were resistant to vancomycin (VAN) by MIC determination,9 with a concentration ranging from 0.25 to 2μg/mL. The majority of MRSA isolates (n=45/73.8%) showed MIC of 1.0μg/mL, following by MIC of 2.0μg/mL (n=14/22.9%). SCCmec types were carried out by multiplex PCR analysis that generated a specific amplification pattern, that includes SCCmec types I, II, III, IV and V, as previously described.10 A total of two (3.3%) MRSA isolates contained SCCmec I, 58 (95.1%) SCCmec IV and one (1.6%) harbored SCCmec II element. Two isolates that containing SCCmec IV, all SCCmec I and II exhibited multidrug resistance (resistance to three or more classes of non-β-lactam antimicrobial drugs). Molecular characterization based on multilocus sequence typing (MLST)11 identified 9 STs previously described among MRSA isolates (ST1, ST5, ST8, ST12, ST30, ST45, ST105, ST435, ST1635). Three new STs (ST3163, ST3164 and ST3165) and three new allelic arrangements were identified and added to the mlst.net database. MLST results showed that most of the isolates (81.9%) belonged to known epidemic lineages. The lineages related with the clones USA800/Pediatric (ST5, SCCmec IV), USA400/MW2/WA-1 (ST1, SCCmec IV), USA1100/OSPC (ST30, SCCmec IV), USA300/USA300-LV (ST8, SCCmec IV), UK EMRSA-3/Cordobes/Chilean (ST5, SCCmec I), USA600/Berlin (ST45, SCCmec IV) and USA100/NYJ (ST5, SCCmec II) were observed in approximately 27.8%, 26.2%, 18%, 3.3%, 3.3%, 1.6% and 1.6% of isolates, respectively. HA-MRSA and CA-MRSA isolates typically found in Brazil, related to USA800 and USA400, respectively, were identified in all hospitals (at least two isolates/hospital). Isolates related to the Oceania Southwest Pacific clone (OSPC/USA1100) were also observed in all hospitals, except H4. Beside, one isolate (new ST3165) belonged to the same clonal complex as OSPC (CC30) was found in the H4 (Table 1). PCR-amplification product12 compatible with the PVL genes lukS and lukF was detected in 11 (18%) of the MRSA blood isolates. Those genes were related to MRSA harboring SCCmec IV. Looking at the epidemic lineage, PVL gene was found in isolates related to OSPC (n=8/72.7%).
Characteristics and identification of 61 MRSA isolates from five hospitals in Rio de Janeiro city.
| ST/CC | n | SCCmec type | Clonality | Resistance profile (n) | Number isolates positives of the lukSF (genes) | H1 (n) | H2 (n) | H3 (n) | H4 (n) | H5 (n) |
|---|---|---|---|---|---|---|---|---|---|---|
| 1/1 | 16 | IV | USA400 | CIP-ERI-CLI-GEN-TET-CLO (1) | – | – | – | 1 | – | |
| CIP-ERI-CLI-GEN-CLO (1) | – | – | – | – | – | 1 | ||||
| CIP-ERI-CLI-CLO-LIN (1) | – | 1 | – | – | – | – | ||||
| CIP-ERI-CLI-GEN (1) | – | – | – | – | 1 | – | ||||
| CIP-ERI-CLI (5) | – | – | 1 | 2 | 1 | 1 | ||||
| CIP-ERI-CLO (1) | – | – | – | – | – | – | ||||
| ERI-TET (1) | – | 1 | – | – | – | 1 | ||||
| TET-CLO (1) | – | – | – | 1 | – | – | ||||
| CIP-ERI (1) | – | – | 1 | – | – | – | ||||
| CIP (1) | – | – | – | 1 | – | – | ||||
| ERI (1) | – | – | – | 1 | – | – | ||||
| Ø (1) | – | 1 | – | – | – | – | ||||
| 5/5 | 20 | I | UK-EMRSA-3 | SUT-CIP-ERI-CLO (1) | – | – | – | 1 | – | – |
| CIP-ERI-CLI (1) | – | – | – | 1 | – | – | ||||
| II | USA100/NYJ | CIP-ERI-CLI-GEN (1) | – | 1 | – | – | – | – | ||
| IV | USA800 | CIP-ERI-CLI-GEN-TET-CLO (1) | – | – | 1 | – | – | – | ||
| CIP-ERI-CLI-RIF-CLO (1) | – | – | – | – | – | 1 | ||||
| CIP-ERI-CLI-CLO (1) | – | – | – | – | – | 1 | ||||
| CIP-ERI-CLI-GEN (3) | – | 1 | – | – | 1 | 1 | ||||
| CIP-ERI-CLI-RIF (1) | – | – | – | – | – | 1 | ||||
| CIP-ERI-CLO (1) | – | – | – | 1 | – | – | ||||
| CIP-ERI-CLI (3) | – | 2 | 1 | – | – | – | ||||
| ERI-CLI (1) | – | – | 1 | – | – | – | ||||
| ERI (5) | – | 1 | 1 | 1 | 1 | 1 | ||||
| 8/8 | 2 | IV | USA300 | CIP-ERI-CLI (1) | – | – | – | – | 1 | – |
| CIP-ERI (1) | – | – | 1 | – | – | – | ||||
| 12/12 | 1 | IV | – | SUT-CIP-ERI-GEN-RIF-LIN (1) | – | – | – | 1 | – | – |
| 30/30 | 11 | IV | OSPC | CIP-ERI-CLI-CLO (1) | 1 | – | 1 | – | – | – |
| CIP-ERI-CLI (2) | 2 | – | 1 | – | – | 1 | ||||
| ERI-CLI-TET (1) | 1 | – | – | 1 | – | – | ||||
| ERI-CLI (2) | 1 | 1 | – | – | – | 1 | ||||
| CIP-ERI (1) | – | – | – | – | – | 1 | ||||
| Ø (4) | 3 | 2 | 2 | – | – | – | ||||
| 45/45 | 1 | IV | USA600/Berlin | CIP-ERI-CLI-RIF-TET-LIN (1) | – | – | – | – | – | 1 |
| 105/5 | 2 | IV | – | CIP-ERI-CLI (2) | – | – | 1 | 1 | – | – |
| 435/30 | 1 | IV | – | Ø (1) | 1 | – | 1 | – | – | – |
| 1635/5 | 3 | IV | – | Ø (3) | – | – | 1 | 1 | 1 | – |
| 3163a/30 | 1 | IV | – | ERI-CLI (1) | 1 | – | – | 1 | – | – |
| 3164a/30 | 2 | IV | – | SUT-CIP-ERI-GEN-RIF-TET-LIN (1) | – | – | – | 1 | – | – |
| Ø (1) | – | – | – | – | – | 1 | ||||
| 3165a/30 | 1 | IV | – | Ø (1) | 1 | – | – | – | 1 | – |
ST, sequence type obtained by MLST; CC, clonal complex; SCCmec, Staphylococcal cassette chromosome mec; H, hospital; –, not found; a, ST described in this study; CLI, clindamycin; ERI, erythromycin; CIP, ciprofloxacin; CLO, chloramphenicol; GEN, gentamicin; RIF, rifampicin; TET, tetracycline; SUT, trimethoprim-sulfamethoxazole; LIN, linezolid; Ø, isolated with resistance only for cefoxitin; lukSF genes encoding Panton-Valentine leucocidin were detected by a PCR-based method.
In recent years, many authors have reported the presence of typically CA-MRSA strains circulating in healthcare environments,2 including Brazil.13,14 The rising frequency of CA-MRSA has several implications in nosocomial infections. CA-MRSA isolates are recognized as presenting a higher spectrum of susceptible to non-β-lactam antibiotics when compared with HA-MRSA. However, vancomycin remains first-choice antibiotic therapy for serious infections.6 The increase use of this late antimicrobial drug is directly related to the emergence of vancomycin-intermediate (VISA), vancomycin-resistant (VRSA) and varied multidrug-resistant MRSA isolates around the world.15,16 In the present study, resistance to vancomycin was not detected; however a MIC creep phenomenon is suggested by the high percentage of samples showing a vancomycin MIC≥1.0μg/mL.15 The highest MIC detected was 2μg/mL and was observed in 22.9% of the isolates. Several authors have reported that treatment with vancomycin eventually fails among patients with MRSA bloodstream infections which presented a vancomycin MIC>1μg/mL.17 Recently, a VRSA strain was isolated from a blood culture in a Hospital in São Paulo city, Brazil, in which the patient had previously been treated with vancomycin.7 A report from Rossi and colleagues7 emphasizes the value of effective surveillance on the MIC levels of vancomycin, as well as the importance of quick response by the clinical microbiology laboratory informing the medical team on the specific vancomycin MIC. Due to the inclusion criteria used in this study, most of the MRSA isolates exhibited SCCmec type IV (95.1%). Differently, Caiaffa-Filho18 studying MRSA blood isolates from Brazilian patients, showed a high frequency of MRSA carrying SCCmec type II. The largest cassettes (I, II and III) enhance the survival abilities of MRSA when in hospital settings. However, it is believed that the smaller cassettes (particularly the IV) promote evolutionary advantages in spreading by horizontal transfer of this element, enabling the increase of CA-MRSA in hospitals.19 Thus CA-MRSA SCCmec IV frequently show competitive advantages compared with multidrug-resistant strains (HA-MRSA).20 Several factors seem to contribute to these adaptive advantages of CA-MRSA isolates, such as the toxin PVL production.5,19 The presence of PVL genes among the isolates characterized as CA-MRSA have been uneven, with percentages of positivity varying from 27 to 75%.19,21 In this study, 11 (18%) isolates were positive to PVL genes and all contained SCCmec IV.
Non multidrug-resistant MRSA isolates carrying SCCmec IV have already been identified in Brazilian hospitals, with patterns similar to USA100, USA300, USA400, USA800 and USA1100 clones,4,13,14,22,23 however, two isolates, in this study related to USA1100 and USA400, showed multidrug resistance profiles.
The emergence of MRSA clones related to USA400 and USA800 was previously described in Rio de Janeiro in only two hospitals,22 and more recently, MRSA related to USA400, isolated from blood, was described associated with infective endocarditis in Rio de Janeiro.24 This multicenter data indicate that these MRSA lineages are still circulating in the Brazilian healthcare settings.
In Brazil the most common lineage of CA-MRSA is related to OSPC, especially causing community associated infections.13,23,25 In this study, eleven isolates (18%) were related to this clone. The presence of isolates related to OSPC clone has already been described in hospitalized patients in two hospitals in Rio de Janeiro,14,22 however, few studies described severe infections by these CA-MRSA from blood,14,26 while in the other case the OSPC related isolate was collected from wound.22 Here, OSPC related isolates were observed in most of the studied hospitals, though, if one considers the clonal complex (CC30), then, we can infer the spread of these MRSA genetically related to OSPC clone in all hospitals. In Uruguay, a Brazilian neighboring country, replacement of HA-MRSA by CA-MRSA in hospital settings have been described, where the most common CA-MRSA isolates recovered from invasive and superficial infection diseases are related to the OSPC clone.20
In the present study, isolates related to USA300, UK EMRSA-3 and USA100 were rarely observed. However, a USA300-related isolate has also been identified in one hospital in Rio de Janeiro from a pneumonia case.13
MRSA belonging to the lineage USA100/ST5-MRSA-II was observed just in one hospital. This lineage has also been observed in blood samples in Brazil, highlighting that one of these reports describes two isolates of S. aureus resistant to daptomycin.27 In Brazil, to the best of our knowledge, there are few reports of USA600/ST45-MRSA-IV isolates, which were collected from skin infection case and from catheter.23,28
The epidemic clone UK MRSA-3 (ST5-MRSA-I) traditionally classified as HA-MRSA is distributed in Europe and Latin America, including Brazil.3,29,30 Using the phenotypic criteria established in this screening, we could identify an isolate related to this clone in our collection of MRSA blood. Recently our group described the presence of this clone in respiratory secretions of cystic fibrosis patients in Rio de Janeiro.1 Until now, there is no report of UK-MRSA-3 in bloodstream infections in Brazil. In conclusion, a number of different international clones, displaying lower level of antimicrobial resistance to non-β lactams, are circulating in different Rio de Janeiro hospitals causing bloodstream infections; including MRSA lineages that are not frequently observed in our country. Thus, we recommend a surveillance including a larger number of MRSA blood isolates, from different Rio de Janeiro hospitals, in order to expose the real picture of the antimicrobial susceptibility profiles found among MRSA isolates, which can significantly vary depending on the dominant lineages present in the studied hospitals.
Conflicts of interestThe authors declare that there is no conflict of interest.
We thank the personnel at the microbiology laboratories of the participant hospitals for their generous collaboration in sending us the MRSA blood isolates. This work was supported by the Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro.