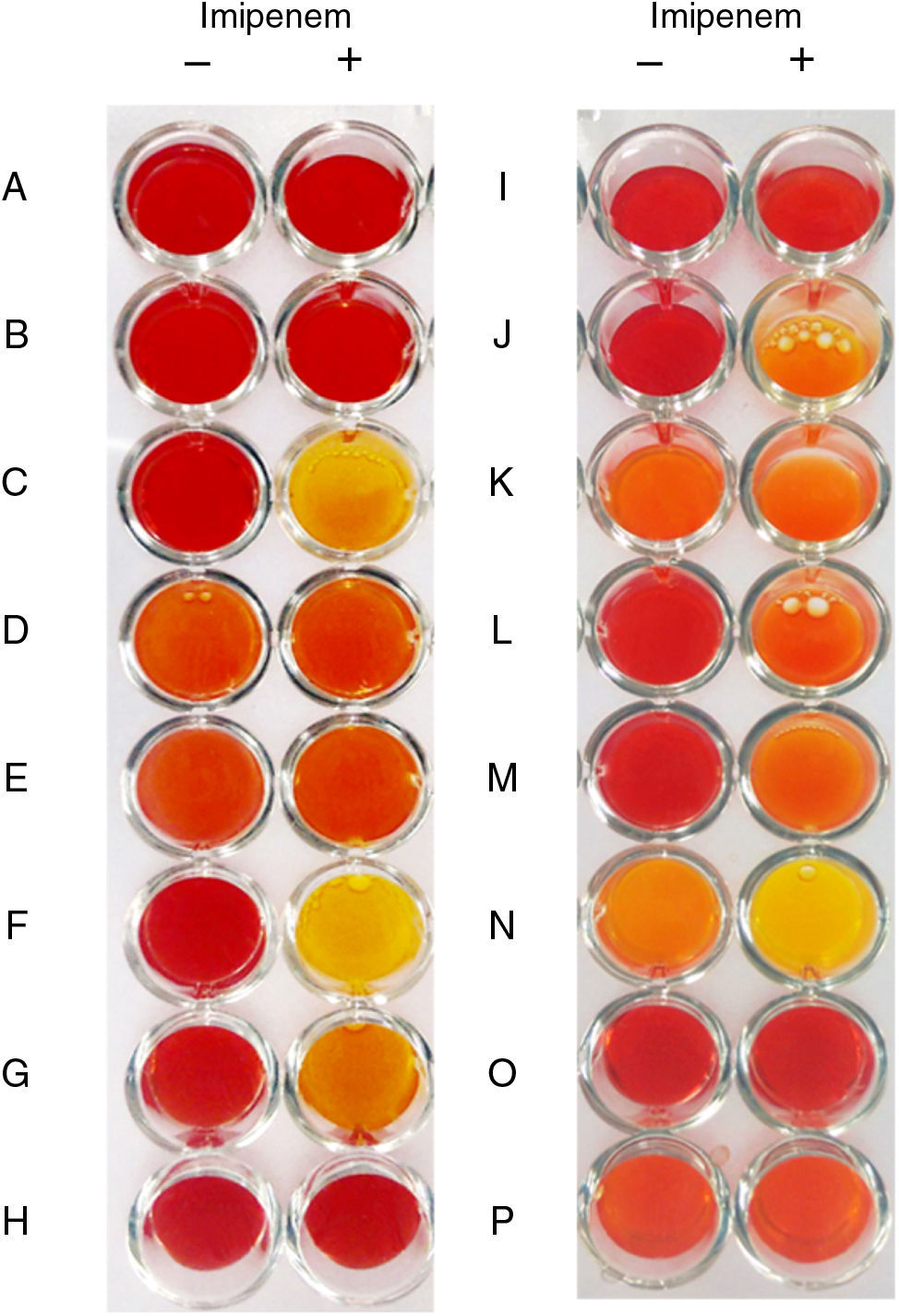
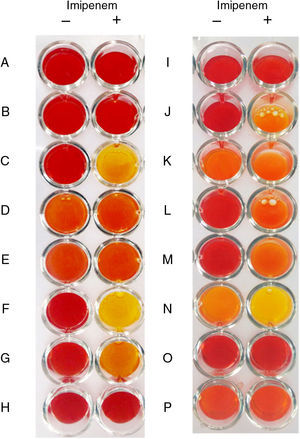
The modified Carba NP test presented here may be a valuable tool for laboratories interested in investigating a large number of carbapenemase-producing bacteria in a less-costly way. The test was evaluated against 48 carbapenemase-producing and carbapenemase-non-producing gram-negative bacteria. No false–positive results were obtained, but false-negative results were observed with OXA-23- and GES-carbapenemase-producing isolates. Aeromonas sp. are not testable by Modified Carba NP.
The spread of carbapenemase-producing isolates in hospital settings is a major public health concern. Early detection of carbapenemase producers is essential to assure adequate therapy and favorable outcomes.1–5 Carba NP test emerged as a useful alternative to detect carbapenemase production in Enterobacteriaceae, Pseudomonas spp. and Acinetobacter spp.,2,6,7 as recommended by the Clinical and Laboratory Standards Institute (CLSI).8 The test is based on acidification of phenol red when imipenem is hydrolyzed, evidenced by the color change of the test solution from red to yellow. Carba NP test advantages over a number of other phenotypic tests include speed in providing results, simplicity of execution, objectiveness in interpretation and increased sensitivity and specificity.1,2,9,10 On the other hand, sample processing may become expensive and time consuming if a large number of isolates are tested. Here we propose modifications to make the test faster and less expensive. Modifications included the omission of the centrifugation step, cell disruption using bath sonication and the use of imipenem/cilastatin as the substrate.
We studied 48 isolates, including negative controls and Enterobacteriaceae, Pseudomonas aeruginosa, Acinetobacter baumannii and Aeromonas spp. producing class A, B or D carbapenemases (Table 1). Bacterial strains were cultivated onto Trypticase soy agar (TSA) (Difco Laboratories) at 37°C for 18–24h. A 10μL calibrated loopfull of the test strain was inoculated into 500μL Tris–HCl (20mM–pH 7.5) (Invitrogen). The suspension was subjected to vortex homogenization and bath sonication for 30min (BRANSONIC ULTRASONIC CLEANER, 47kHz±6%, 60W) and preserved on ice. Then, 30μL of the cell extract was mixed with 100μL of phenol red (Isofar) containing 0.1mM ZnSO4 (Merck) (imipenem −) and 100μL phenol red containing 0.1mM ZnSO4 and 6mg/mL imipenem/cilastatin (Merck) (imipenem +). The mixtures were incubated at 37°C for 2h. Tests were performed in duplicate for all isolates. The color change of the imipenem-containing vial from red to yellow or orange indicated a positive result. Three independent observers recorded the results with no discordant readings.
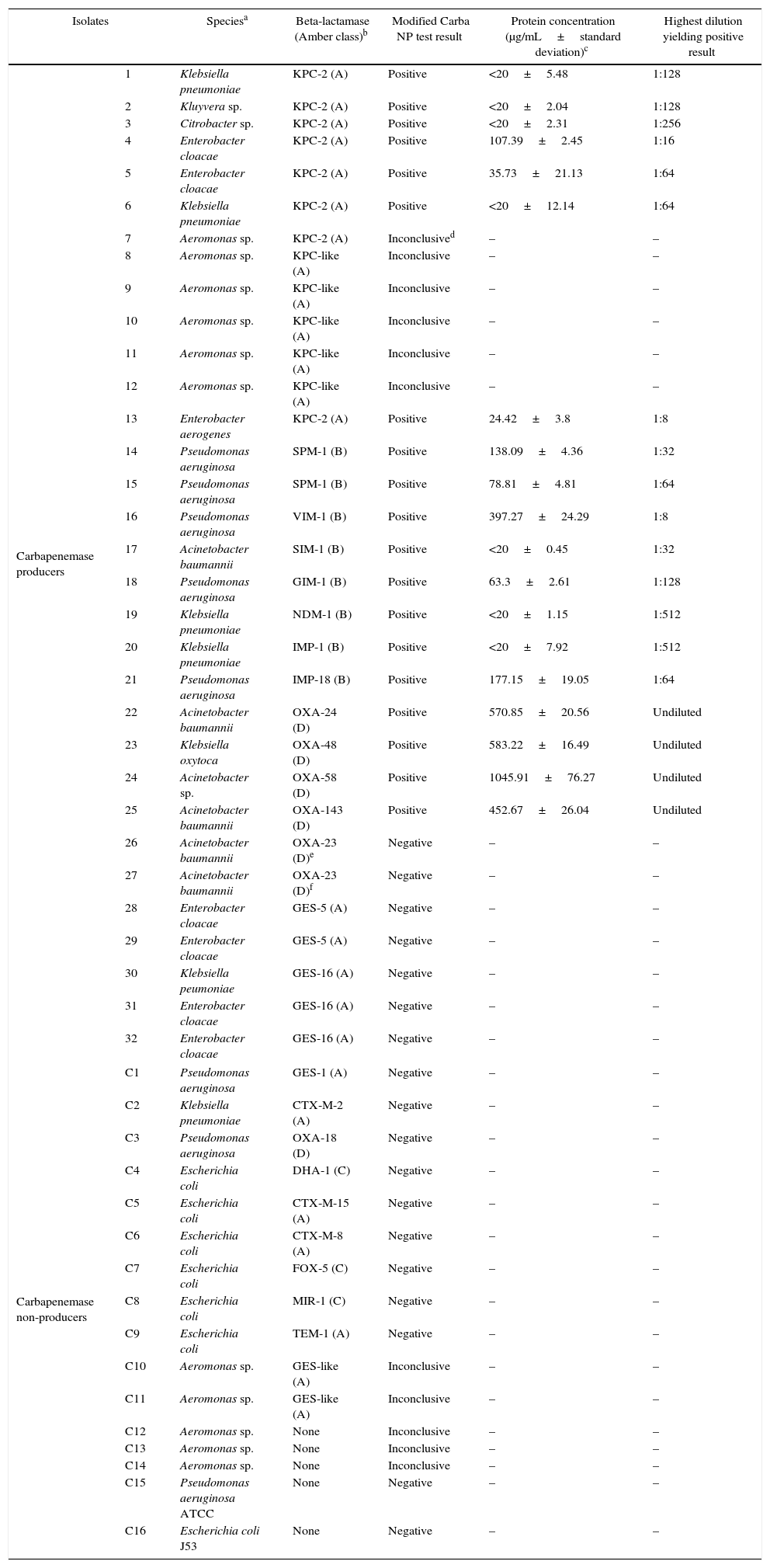
Isolates tested, modified Carba NP results and protein concentration.
| Isolates | Speciesa | Beta-lactamase (Amber class)b | Modified Carba NP test result | Protein concentration (μg/mL±standard deviation)c | Highest dilution yielding positive result | |
|---|---|---|---|---|---|---|
| Carbapenemase producers | 1 | Klebsiella pneumoniae | KPC-2 (A) | Positive | <20±5.48 | 1:128 |
| 2 | Kluyvera sp. | KPC-2 (A) | Positive | <20±2.04 | 1:128 | |
| 3 | Citrobacter sp. | KPC-2 (A) | Positive | <20±2.31 | 1:256 | |
| 4 | Enterobacter cloacae | KPC-2 (A) | Positive | 107.39±2.45 | 1:16 | |
| 5 | Enterobacter cloacae | KPC-2 (A) | Positive | 35.73±21.13 | 1:64 | |
| 6 | Klebsiella pneumoniae | KPC-2 (A) | Positive | <20±12.14 | 1:64 | |
| 7 | Aeromonas sp. | KPC-2 (A) | Inconclusived | – | – | |
| 8 | Aeromonas sp. | KPC-like (A) | Inconclusive | – | – | |
| 9 | Aeromonas sp. | KPC-like (A) | Inconclusive | – | – | |
| 10 | Aeromonas sp. | KPC-like (A) | Inconclusive | – | – | |
| 11 | Aeromonas sp. | KPC-like (A) | Inconclusive | – | – | |
| 12 | Aeromonas sp. | KPC-like (A) | Inconclusive | – | – | |
| 13 | Enterobacter aerogenes | KPC-2 (A) | Positive | 24.42±3.8 | 1:8 | |
| 14 | Pseudomonas aeruginosa | SPM-1 (B) | Positive | 138.09±4.36 | 1:32 | |
| 15 | Pseudomonas aeruginosa | SPM-1 (B) | Positive | 78.81±4.81 | 1:64 | |
| 16 | Pseudomonas aeruginosa | VIM-1 (B) | Positive | 397.27±24.29 | 1:8 | |
| 17 | Acinetobacter baumannii | SIM-1 (B) | Positive | <20±0.45 | 1:32 | |
| 18 | Pseudomonas aeruginosa | GIM-1 (B) | Positive | 63.3±2.61 | 1:128 | |
| 19 | Klebsiella pneumoniae | NDM-1 (B) | Positive | <20±1.15 | 1:512 | |
| 20 | Klebsiella pneumoniae | IMP-1 (B) | Positive | <20±7.92 | 1:512 | |
| 21 | Pseudomonas aeruginosa | IMP-18 (B) | Positive | 177.15±19.05 | 1:64 | |
| 22 | Acinetobacter baumannii | OXA-24 (D) | Positive | 570.85±20.56 | Undiluted | |
| 23 | Klebsiella oxytoca | OXA-48 (D) | Positive | 583.22±16.49 | Undiluted | |
| 24 | Acinetobacter sp. | OXA-58 (D) | Positive | 1045.91±76.27 | Undiluted | |
| 25 | Acinetobacter baumannii | OXA-143 (D) | Positive | 452.67±26.04 | Undiluted | |
| 26 | Acinetobacter baumannii | OXA-23 (D)e | Negative | – | – | |
| 27 | Acinetobacter baumannii | OXA-23 (D)f | Negative | – | – | |
| 28 | Enterobacter cloacae | GES-5 (A) | Negative | – | – | |
| 29 | Enterobacter cloacae | GES-5 (A) | Negative | – | – | |
| 30 | Klebsiella peumoniae | GES-16 (A) | Negative | – | – | |
| 31 | Enterobacter cloacae | GES-16 (A) | Negative | – | – | |
| 32 | Enterobacter cloacae | GES-16 (A) | Negative | – | – | |
| Carbapenemase non-producers | C1 | Pseudomonas aeruginosa | GES-1 (A) | Negative | – | – |
| C2 | Klebsiella pneumoniae | CTX-M-2 (A) | Negative | – | – | |
| C3 | Pseudomonas aeruginosa | OXA-18 (D) | Negative | – | – | |
| C4 | Escherichia coli | DHA-1 (C) | Negative | – | – | |
| C5 | Escherichia coli | CTX-M-15 (A) | Negative | – | – | |
| C6 | Escherichia coli | CTX-M-8 (A) | Negative | – | – | |
| C7 | Escherichia coli | FOX-5 (C) | Negative | – | – | |
| C8 | Escherichia coli | MIR-1 (C) | Negative | – | – | |
| C9 | Escherichia coli | TEM-1 (A) | Negative | – | – | |
| C10 | Aeromonas sp. | GES-like (A) | Inconclusive | – | – | |
| C11 | Aeromonas sp. | GES-like (A) | Inconclusive | – | – | |
| C12 | Aeromonas sp. | None | Inconclusive | – | – | |
| C13 | Aeromonas sp. | None | Inconclusive | – | – | |
| C14 | Aeromonas sp. | None | Inconclusive | – | – | |
| C15 | Pseudomonas aeruginosa ATCC | None | Negative | – | – | |
| C16 | Escherichia coli J53 | None | Negative | – | – | |
To assess the inferior limit of carbapenemase detection by the modified Carba NP proposed, the test was also performed using diluted crude extracts. The last dilution yielding positive result was centrifuged and the supernatant was subjected to total protein quantification, which was performed in triplicate using the Pierce™ BCA Protein Assay Kit (Thermo Scientific) following the manufacturer's recommendations.
Sensibility, specificity, positive and negative predictive values (SN, SP, PPV and NPV, respectively) were calculated, excluding Aeromonas spp. PCR results for carbapenamases were considered the gold standard. SN, SP, PPV, and NPV were calculated with the formulas a/(a+c), d/(b+d), a/(a+b) and d/(c+d), respectively.
Most carbapenemase-producing isolates showed the expected positive result, indicating that the modification in the extraction protocol did not jeopardize the sensitivity of the test (Table 1). Protein concentrations at the most diluted extract showing carbapenemase activity varied from <20μg/mL to 1045.92μg/mL (Table 1).
All KPC producing isolates yielded positive results except for Aeromonas sp. (Table 1) which showed inconclusive results in repeated tests irrespective of the beta-lactamase produced, as the red-to-yellow color change was observed in solutions imipenem − and + (Fig. 1). This finding was related to the acidic nature of the crude extract assessed in repeated assays (pH=5.5). Although Aeromonas spp. producing acquired carbapenemases are not common causes of multidrug-resistant infections, microbiologists should be aware that Carba NP test might not be suitable to investigate carbapenemase production in these bacteria. All metalo-β-lactamase (MBL) producers showed positive results at the modified Carba NP test, as expected. Of notice, MBL-producing P. aeruginosa required increased protein levels in crude extracts to generate positive results, likely due to difficulty in disrupting the cell wall of such isolates. In agreement with previous work,2,11,12 all GES-like carbapenemase producing isolates showed negative results. Most class D carbapenemase producing isolates showed positive results in Modified Carba NP test, except those producing OXA-23 (Table 1). Diluted cell extracts, however, showed negative results, which is consistent with the decreased imipenem catalytic activity presented by oxacilinases compared to other carbapenemases. Isolates carrying blaOXA-23 showed negative results, regardless of the presence or absence of ISAba1 upstream this gene. The SN, SP, PPV, and NPV for Carba NP modified were 73.1, 100, 100 and 61.1%, respectively. Positive results were observed at different times for different carbapenemases (ranging from 5min for NDM and KPC to 2h for OXA type).
Representative results of the modified Carba NP test. Non-carbapenemase producers (A, B, H and K), carbapenemase producers (C, F, G, I, J, L, M, and O), and Aeromonas spp. isolates (D, E, K, N and P) with negative control solutions (−) and test solution (+). (A) E. coli J-53; (B) P. aeruginosa ATCC 25922; (C) KPC-2-producing E. cloacae; (D) KPC-like-producing Aeromonas sp. (E) KPC-like-producing Aeromonas sp. (F) NDM-1-producing K. pneumoniae; (G) OXA-48-producing K. oxytoca; (H) CTX-M_15-producing E. coli; (I) OXA-23-producing A. baumannii; (J) OXA-143-producing A. baumannii; (K) Aeromonas sp. (L) IMP-18-producing P. aeruginosa; (M) SPM-1-producing P. aeruginosa; (N) KPC-like-producing Aeromonas sp. (O) GES-16-producing E. cloacae; (P) GES-like-producing Aeromonas sp.
Noteworthy, the Modified Carba NP test gave indistinguishable results when performed using cell extracts obtained by probe sonication (data not shown). Despite the fact that the equipment for bath sonication is cheaper than the probe-based, it also enables processing a large number of isolates concomitantly and avoids excessive manipulation of potential carbapenemase producers in high inoculums, protecting against environment contamination.
Although other studies have made different changes in Carba NP,13–15 the Modified Carba NP test presented here may be a valuable tool for laboratories interested in investigating several carbapenemase-producing bacteria with decreased cost. Although these modifications involve the acquisition of a sonication apparatus, its initial cost is counterbalanced by the ability to process several isolates concomitantly and by the elimination of the lysis buffer, which is especially attractive for laboratories that must import this expensive reagent. We also reinforce that imipenem/cilastatin available in hospital pharmacies may serve as the substrate for the Modified Carba NP test, representing an off-label use of this medication in institutions where this practice is allowed.
Conflicts of interestThe authors declare no conflicts of interest.
We are grateful to Ana Cristina Gales, Laurent Poirel and Marise Dutra Asensi for providing positive controls employed in this study. This work was supported by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ).