Actinobacteria occur in many environments and have the capacity to produce secondary metabolites with antibiotic potential. Identification and taxonomy of actinobacteria that produce antimicrobial substances is essential for the screening of new compounds, and sequencing of the 16S region of ribosomal DNA (rDNA), which is conserved and present in all bacteria, is an important method of identification. Melanized fungi are free-living organisms, which can also be pathogens of clinical importance. This work aimed to evaluate growth inhibition of melanized fungi by actinobacteria and to identify the latter to the species level. In this study, antimicrobial activity of 13 actinobacterial isolates from the genus Streptomyces was evaluated against seven melanized fungi of the genera Exophiala, Cladosporium, and Rhinocladiella. In all tests, all actinobacterial isolates showed inhibitory activity against all isolates of melanized fungi, and only one actinobacterial isolate had less efficient inhibitory activity. The 16S rDNA region of five previously unidentified actinobacterial isolates from Ilha do Mel, Paraná, Brazil, was sequenced; four of the isolates were identified as Streptomyces globisporus subsp. globisporus, and one isolate was identified as Streptomyces aureus. This work highlights the potential of actinobacteria with antifungal activity and their role in the pursuit of novel antimicrobial substances.
Actinobacteria occur in various environments1 and have the capacity to produce extracellular enzymes and secondary metabolites with antibiotic properties,2 thus showing significant biotechnological and therapeutic potential.3 Actinobacteria from intertidal regions produce unique metabolites and carry out unique physiological processes due to extreme environmental conditions, such as salinity, temperature, and humidity.4 Actinobacteria from the intertidal region of Ilha do Mel, Paraná, Brazil, have already shown promising results in inhibition of pathogenic organisms and production of substances with antimicrobial potential.5 Other studies have found antibacterial and antifungal properties in extracts from actinobacteria isolated from forest soil,6 highlighting the importance of studying antimicrobial potential of secondary metabolites produced by actinobacteria.
Melanized fungi, which are dark-colored organisms due to the presence of melanin in their cell wall,7 can be saprotrophic or pathogenic to humans, vertebrates, or plants. Diseases caused by melanized fungi are known as eumycetoma, chromoblastomycosis, and phaeohyphomycosis. These fungi usually belong to the genera Exophiala, Cladosporium, and Rhinocladiella. The treatment is usually clinical or surgical, and the drugs itraconazole and ketoconazole are frequently used to treat these infections.8
Identification and taxonomy of actinobacteria are very important for the research of new compounds, providing information about the relations between organisms and about their potential secondary metabolites.9 An important method for identification of actinobacteria is the sequencing of the 16S region of their ribosomal DNA (rDNA). This region is conserved and present in all bacteria.10 Comparison of sequences from unidentified isolates with those already known allows the construction of phylogenetic trees and identification of organisms.11 The aim of this work was to evaluate inhibitory activity of actinobacteria against melanized fungi and identify the actinobacterial isolates to the species level.
Materials and methodsMicrobial strainsIn this study, 13 actinobacterial strains (Table 1) previously isolated from marine sediments5 and seven strains of melanized fungi (Table 1) previously isolated from dialysis water of various dialysis units in Curitiba, Brazil,12 were used in inhibitory activity tests.
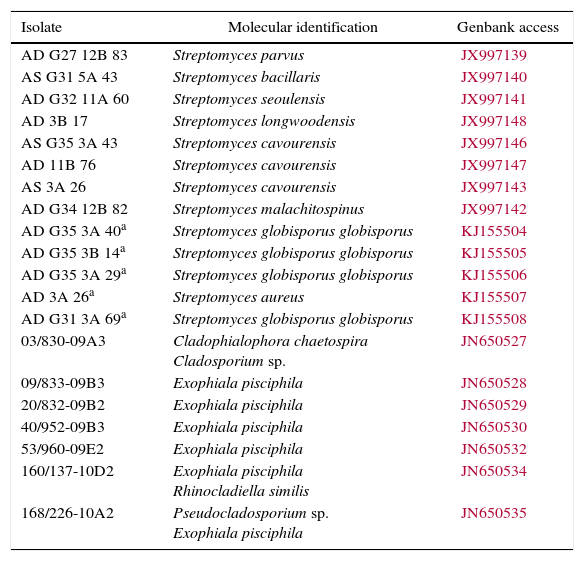
| Isolate | Molecular identification | Genbank access |
|---|---|---|
| AD G27 12B 83 | Streptomyces parvus | JX997139 |
| AS G31 5A 43 | Streptomyces bacillaris | JX997140 |
| AD G32 11A 60 | Streptomyces seoulensis | JX997141 |
| AD 3B 17 | Streptomyces longwoodensis | JX997148 |
| AS G35 3A 43 | Streptomyces cavourensis | JX997146 |
| AD 11B 76 | Streptomyces cavourensis | JX997147 |
| AS 3A 26 | Streptomyces cavourensis | JX997143 |
| AD G34 12B 82 | Streptomyces malachitospinus | JX997142 |
| AD G35 3A 40a | Streptomyces globisporus globisporus | KJ155504 |
| AD G35 3B 14a | Streptomyces globisporus globisporus | KJ155505 |
| AD G35 3A 29a | Streptomyces globisporus globisporus | KJ155506 |
| AD 3A 26a | Streptomyces aureus | KJ155507 |
| AD G31 3A 69a | Streptomyces globisporus globisporus | KJ155508 |
| 03/830-09A3 | Cladophialophora chaetospira Cladosporium sp. | JN650527 |
| 09/833-09B3 | Exophiala pisciphila | JN650528 |
| 20/832-09B2 | Exophiala pisciphila | JN650529 |
| 40/952-09B3 | Exophiala pisciphila | JN650530 |
| 53/960-09E2 | Exophiala pisciphila | JN650532 |
| 160/137-10D2 | Exophiala pisciphila Rhinocladiella similis | JN650534 |
| 168/226-10A2 | Pseudocladosporium sp. Exophiala pisciphila | JN650535 |
All organisms were stored in the biological collection of LabMicro, Universidade Federal do Paraná, Curitiba, Brazil.
Inhibition testsInhibitory activity of Streptomyces spp. against the melanized fungi belonging to the genera Exophiala, Rhinocladiella, and Cladosporium was evaluated using inhibition tests.13
The inhibition tests consisted of spreading a saline solution with 3×108 actinobacterial cells per milliliter, according to the McFarland turbidity scale, on Sabouraud agar medium in a Petri dish using a Drigalski spatula. A small block with a diameter of 6mm was removed from the center of the dish and replaced with another one containing a fungal culture grown for 10 days at 27°C on Sabouraud agar. The control consisted of a Petri dish containing the fungal culture alone. All tests were performed in triplicate.
The growth diameter of the fungal isolates was measured after 7 and 14 days of incubation at 27°C on Sabouraud agar. The growth of the fungus in Petri dishes that contained actinobacteria was then compared to the growth of the control samples using statistical analysis.
The data were transformed using log (x+2) and analyzed using analysis of variance and Tukey's test at 5% probability. In addition, a factorial experiment (1×1) was performed. All statistical tests were performed using the ASSISTAT 7.6 software.14
16S rDNA sequencingAll organisms have been previously identified morphologically as belonging to the genus Streptomyces, and eight out of the 13 strains tested have been previously identified using molecular methods (Table 1).5 The remaining strains, AD G35 3A 29, AD G35 3A 40, AD G35 3B 14, AD 3A 26, and AD G31 13A 69, were identified in this work using 16S rDNA sequencing. Actinobacterial isolates were grown for three days at 27°C in Czapek–Dox medium. DNA was extracted as previously described16 and amplified using the primers 9F (5′ GAGTTTGATCCTGGCTCAG 3′) and Sm5R (5′ GAACTGAGACCGGCTTTTTGA 3′). Denaturation of DNA was performed at 95°C for 5min, followed by 30 cycles of 45s at 94°C, 45s at 65°C, and 1min at 72°C, and a final extension of 10min at 72°C.15 Polymerase chain reaction products were purified and sequenced using an ABI 3130 sequencer (Applied Biosystems).16
The sequences were then analyzed using the Staden 1.6 software17 and aligned using the MEGA 4.0 software.18 The sequences were then compared to those deposited to the National Center for Biotechnology Information database using the BLAST algorithm.19
Results and discussionInhibition testsThe 13 actinobacterial isolates were screened for inhibitory activity against the seven isolates of melanized fungi (Table 1). In all tests, statistically significant inhibitory activity was observed, and the F-value was also highly significant (p<0.01).
To identify the actinobacterial isolate with the greatest inhibition potential, a factorial experiment (1×1) was performed. The F-value was highly significant (p<0.01), demonstrating a similar inhibition potential of 12 out of the 13 isolates tested. The only isolate that showed a lower inhibitory efficiency was AD G31 13A 69 (Streptomyces globisporus subsp. globisporus). Fungal strains 03/830-09A3 (Cladosporium sp.) and 40/952-09B3 (Exophiala pisciphila) were more efficiently inhibited by the actinobacteria assayed than the other fungal isolates.
The 13 actinobacterial isolates tested in the present work had already shown inhibitory activity against other pathogenic microorganisms in a previous study.5 Among the 116 actinobacteria isolated by the author from intertidal regions of Ilha do Mel, Paraná, Brazil, 68% showed activity against the pathogenic strains Staphylococcus aureus ATCC 25923, Candida albicans ATCC 10231, Escherichia coli ATCC 25922, and Pseudomonas aeruginosa ATCC 27853. This study highlighted the inhibitory activity of these actinobacteria and their potential to produce antimicrobial and antifungal compounds.
Other authors have also demonstrated inhibitory activity of actinobacteria from the genus Streptomyces against other microorganisms. Strains of Streptomyces spp. were tested against the phytopathogenic fungi Phytophthora parasitica, Guignardia citricarpa, Rhizoctonia solani, Colletotrichum sublineolum, Pythium sp., and Fusarium oxysporum. Most of the actinobacterial strains showed inhibitory activity against the fungi. This activity was often species-related, showing the importance of identifying organisms with antimicrobial potential. The results of this study have also revealed a correlation of fungal inhibition with chitinolytic activity, indicating a role of compounds produced by actinobacteria in antimicrobial activity.20
In another study, inhibitory activity has also been detected in actinobacterial isolates from the genera Streptomyces, Nocardia, Kitasatospora, Amycolatopsis, Rhodococcus, and Gordonia against Bacillus subtilis, E. coli, C. albicans, Xanthomonas campestris, and Mucor racemosus. The results of this study indicated the inhibition potential of actinobacteria from the genus Streptomyces against diverse microorganisms.21
These results highlight the relevance of actinobacteria to the screening for new compounds with antimicrobial activity and their potential to produce antimicrobial and antifungal compounds.
16S rDNA sequencingThe 16S rDNA sequences were edited using the Staden software, and the BLAST algorithm (http://blast.ncbi.nlm.nih.gov) was used to compare the sequences with other sequences previously deposited in the GenBank database. The sequences were aligned to those of type strains using the MEGA 5.2 software. Following the alignment, a dendrogram was constructed using the neighbor-joining method with 1000 bootstrap replicates, the Tamura three-parameter model, and the Gamma (G) distribution (Fig. 1) in the same software.
16S rDNA of the actinobacterial isolates AD G35 3A 29, AD G35 3A 40, AD G35 3B 14, AD 3A 26, and AD G31 13A 69 was sequenced. Four of these isolates were identified as S. globisporus subsp. globisporus, and one was identified as S. aureus.
Other authors have previously isolated S. globisporus from diverse environments, such as soil22 and mangroves.23
S. globisporus has been isolated from soil samples from China, and its secondary metabolites were isolated and evaluated. Compound C-1027 was tested for antimicrobial and antitumor activities and showed antimicrobial activities against most gram-positive bacteria; however, no activity was observed against Mycobacterium sp. or gram-negative bacteria. This compound was also tested against KB carcinoma cells in vitro, and a potent cytotoxicity effect was observed, as well as the ability to inhibit transplantable tumors in mice.22
S. aureus has been found in Thailand soils21 and mangroves in India.23 The antifungal activity of S. aureus has been tested against the phytopathogenic fungus Valsa paulowniae,24 and inhibition of the fungal growth was observed.
Besides antimicrobial activity, S. aureus can also produce metabolites able to degrade pollutants. Strain HP-S-01 was isolated from activated sludge; its capability of degrading soil pollutants was measured, and its metabolite 3-phenoxybenzaldehyde was analyzed. The results showed that 47.9% of β-cypermethrin and 67.0% of 3-phenoxybenzaldehyde were degraded in the absence of additional carbon sources. The study suggested that the HP-S-01 strain of S. aureus can be used for bioremediation; however, more research is needed before its application at a large scale.25
The results of this study led us to conclude that the isolates of Streptomyces spp. from Ilha do Mel, Paraná, Brazil, have a statistically significant potential to inhibit growth of melanized fungi from the genera Exophiala, Cladosporium, and Rhinocladiella. Among the five previously unidentified isolates tested in the present study, four were identified as S. globisporus subsp. globisporus, and one was identified as S. aureus. Further research is necessary to better understand this inhibition potential and its mechanisms. These results also highlighted the importance of actinobacteria in the pursuit of new compounds with antimicrobial potential.
Conflicts of interestThe authors declare no conflicts of interest.