The epidemiology of Helicobacter pylori resistance to antibiotics is poorly documented in Africa and especially in Algeria. The aim of our study was to determine the antibiotic resistance rates, as well as its possible relationship with VacA and CagA virulence markers of isolates from Algerian patients. One hundred and fifty one H. pylori isolate were obtained between 2012 and 2015 from 200 patients with upper abdominal pain. Antimicrobial susceptibility testing was performed for amoxicillin, clarithromycin, metronidazole, ciprofloxacin, rifampicin and tetracycline. Molecular identification of H. pylori and the detection of vacA and cagA genes were performed using specific primers. We found that H. pylori was present in 83.5% of collected biopsies, 54.9% of the samples were cagA positive, 49.67% were vacA s1m1, 18.30% were vacA s1m2 and 25.49% were vacA s2m2. Isolates were characterized by no resistance to amoxicillin (0%), tetracycline (0%), rifampicin (0%), a high rate of resistance to metronidazole (61.1%) and a lower rate of resistance to clarithromycin (22.8%) and ciprofloxacin (16.8%). No statically significant relationship was found between vagA and cagA genotypes and antibiotic resistance results (p>0.5) except for the metronidazole, which had relation with the presence of cagA genotype (p=0.001).
Helicobacter pylori (H. pylori) is a gram negative spiral chapped bacterium, described in 1982 by Marshal and Warren. It specifically infects the gastric mucosa. This discovery had changed all data about gastro-duodenal disorders. H. pylori infects approximately fifty per cent of the world's population, it is an important risk factor in chronic gastritis, peptic ulcer disease, gastric carcinoma and mucosa-associated lymphoid tissue (MALT) lymphoma. It chronically infects most people in the developing countries, and also remains a significant pathogen in industrialized countries.1 In Algeria, its prevalence is average 80% in peptic ulcer.2 Fortunately, many H. pylori associated pathologies can be prevented or cured by the eradication of the bacterium.
Until now, in Algeria eradication of H. pylori is recommended by a triple therapy which includes amoxicillin, clarithromycin or metronidazole combined with proton pump inhibitors (PPI) for 7–10 days.3 It is effective for 79–85% of patients but after the publication of the Maastricht V, Toronto and Spanish Consensus about management of H. pylori infection and its treatment in adults,4,5 recommendations for eradication, therapy in Algeria should be changed, at least in length in order to gain an eradication >=90% of treated patients.
Eradication efforts fail for a significant proportion of patients in Algeria for several reasons, including bacterial resistance to clarithromycin. Several studies have demonstrated that primary resistance to clarithromycin is a major factor for therapeutic failure; the rate of resistance to clarithromycin is significantly increasing and the high rate of the resistance to metronidazole leads to avoid the combination of these two antibiotics in the treatment regimens.3 In another way, results of some studies have suggested that the eradication rate in patients with gastritis is lower than in those with peptic ulcer diseases.6 Since the worldwide increase of the drug resistance rates represents a problem of relevance, some researches have been conducted on antibiotic resistance relationship with bacterial genetic factors such as the cytotoxin associated gene A (cagA) and vaculating cytotoxin gene A (vacA).7,8
Because of the limited data about the resistance of H. pylori to antibiotics in Algeria, the aim of this study was to investigate the resistance rates of amoxicillin, clarithromycin, metronidazole, tetracycline, ciprofloxacin and rifampicin, and to study the relation between H. pylori genotypes and the resistance to antibiotics.
Materials and methods patientsFrom 2012 to 2015, four Hospitals were involved in the present study: Mustapha Pacha University Hospital (Algiers), Halouche clinic of Gastroenterology (Chlef), First November 1954 University Hospital of Oran and Sidi Belabess University Hospital. Two hundred patients with upper abdominal pain from four different regions in Algeria (Algiers, Chlef, Oran and Sidi Belabess), were enrolled in the present study (91 males and 109 females). The patients were Algerian citizens aging from 18 to 86 years and provided written informed consent before endoscopy. Patients either receiving proton pump inhibitory drugs or have been treated with antimicrobials were excluded from the study. All subjects underwent endoscopy to obtain two antrum biopsies from each patient. The first was used for screening of H. pylori positive specimens by a rapid urease test (RUT). While the second piece was placed in 1mL of sterile phosphate buffer saline solution and was transported immediately for the isolation of H. pylori. This study protocol was approved by the Algerian national ethics committee.
Bacterial cultureH. pylori was isolated from grinded gastric biopsy samples on brain heart infusion (BHI) agar (bioMérieux) supplemented with 10% defibrinated horse blood, 0.4% IsoVitaleX, 5mg/L of trimethoprim, 5mg/L of cefsulodin, 10mg/L of vancomycin, and 8mg/L of amphotericin B. The plates were incubated at 37°C under microaerobic conditions for 7 days or more. H. pylori was identified by colony aspect, microscopic morphology, positive urease, catalase and oxidase tests. Strains were stored at −80°C in BHI broth with 20% glycerol.9,10
Antimicrobial susceptibility testingAntibiotic susceptibility was determined using the E-test and agar dilution methods as recommended by EUCAST. The tested antibiotics were amoxicillin (AMX), clarithromycin (CLA), ciprofloxacin (CIP), metronidazole (MTZ), rifampicin (RIF) and tetracycline (TET). The breakpoints used to classify isolates as susceptible or resistant according to the MIC value according to EUCAST: the isolate is resistant to AMX if MIC >0.12mg/L, resistant to CLA if MIC >0.5mg/L, resistant to CIP if MIC >1mg/L; resistant to MTZ if MIC >8mg/L; resistant to RIF if MIC >1mg/L and resistant to TET if MIC >1mg/L.11 Mueller–Hinton agar supplemented with 7% defibrinated horse blood (bioMérieux) was used as the culture medium. An H. pylori culture suspension with a turbidity equivalent of a 3.0 McFarland standard was used to inoculate the plates containing serial dilutions of antibiotics: CLA, 0.016–256mg/L; AMX, 0.016–32mg/L; MTZ, 0.016–256mg/L; TET, CIP and RIF, 0.016–32mg/L (bioMérieux), or plates without antibiotics onto E-test strips of each antibiotic tested were applied (bioMérieux). The plates were incubated at 37°C for 3 days under microaerophilic conditions.11
DNA extraction and purificationTotal genomic DNA was extracted using commercial kits: Magazorb DNA mini-prep kit (Promega) for biopsies and Wizard Genomic DNA Purification Kit (Promega) for strains according to the manufacturer's guidelines. Genomic DNA from biopsies and strains was stored at −20°C.
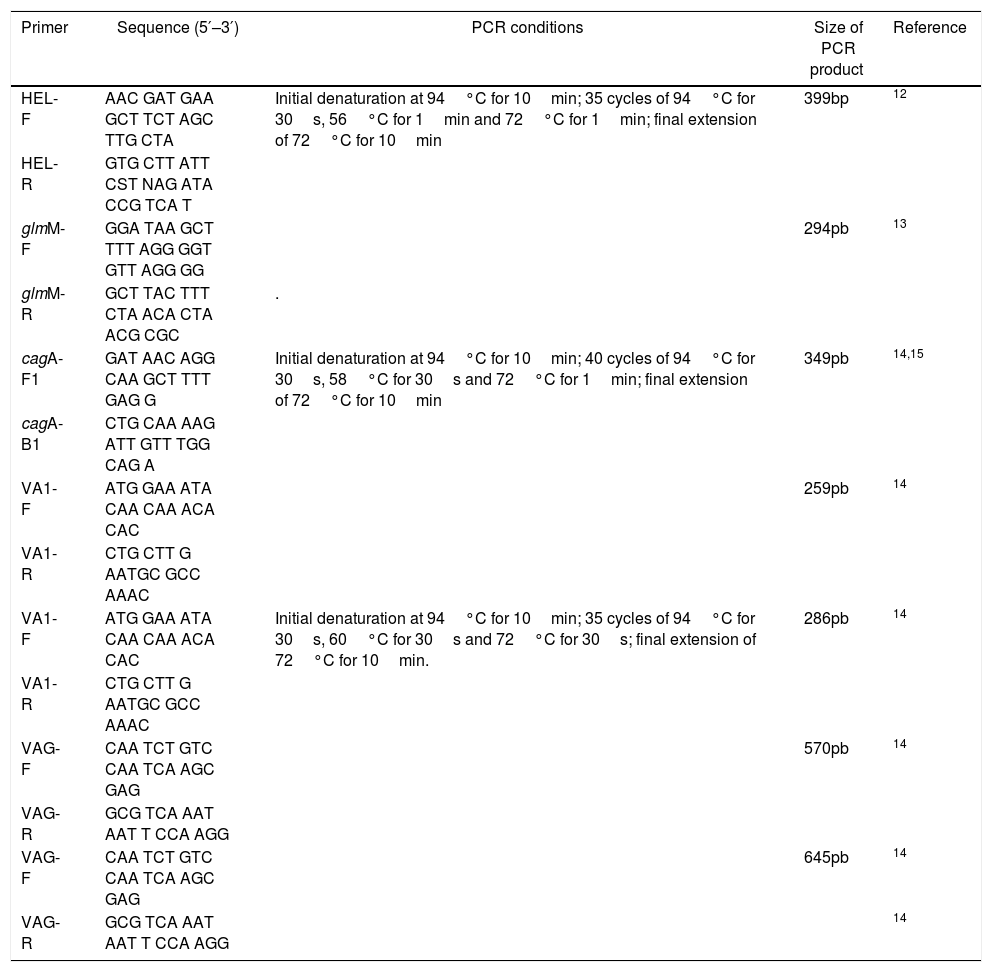
Molecular detection of H. pyloriMolecular detection of H. pylori in biopsy DNA and strain identification were performed using the primers in Table 1. The 25μL reaction mixture consisted of 1× PCR buffer, 1.5mM magnesium chloride, 200μM of each dNTP, 20pmol of each primer and 1U hot start Taq DNA polymerase (Promega). Amplification was carried out in an Eppendorf Mastercycler gradient using the cycling parameters in Table 1. The PCR products were separated on a 2% agarose gel and 100bp ladder was used as DNA molecular weight standard.12,13
Primers used for polymerase chain reaction analysis.
| Primer | Sequence (5′–3′) | PCR conditions | Size of PCR product | Reference |
|---|---|---|---|---|
| HEL-F | AAC GAT GAA GCT TCT AGC TTG CTA | Initial denaturation at 94°C for 10min; 35 cycles of 94°C for 30s, 56°C for 1min and 72°C for 1min; final extension of 72°C for 10min | 399bp | 12 |
| HEL-R | GTG CTT ATT CST NAG ATA CCG TCA T | |||
| glmM-F | GGA TAA GCT TTT AGG GGT GTT AGG GG | 294pb | 13 | |
| glmM-R | GCT TAC TTT CTA ACA CTA ACG CGC | . | ||
| cagA-F1 | GAT AAC AGG CAA GCT TTT GAG G | Initial denaturation at 94°C for 10min; 40 cycles of 94°C for 30s, 58°C for 30s and 72°C for 1min; final extension of 72°C for 10min | 349pb | 14,15 |
| cagA-B1 | CTG CAA AAG ATT GTT TGG CAG A | |||
| VA1-F | ATG GAA ATA CAA CAA ACA CAC | 259pb | 14 | |
| VA1-R | CTG CTT G AATGC GCC AAAC | |||
| VA1-F | ATG GAA ATA CAA CAA ACA CAC | Initial denaturation at 94°C for 10min; 35 cycles of 94°C for 30s, 60°C for 30s and 72°C for 30s; final extension of 72°C for 10min. | 286pb | 14 |
| VA1-R | CTG CTT G AATGC GCC AAAC | |||
| VAG-F | CAA TCT GTC CAA TCA AGC GAG | 570pb | 14 | |
| VAG-R | GCG TCA AAT AAT T CCA AGG | |||
| VAG-F | CAA TCT GTC CAA TCA AGC GAG | 645pb | 14 | |
| VAG-R | GCG TCA AAT AAT T CCA AGG | 14 |
The presence of the genotype cagA was determined by PCR using primers in Table 1. DNA sample from H. pylori J99 strain was used as positive control and sterile distilled water was used as a negative control. The 10μL reaction mixture consisted of 1× PCR buffer, 1.5mM magnesium chloride, 200μM dNTP, 0.5pmol of each primer, 0.5U hot start taq polymerase (Promega) and 2μL of template DNA. Amplification was carried out in an Eppendorf Mastercycler gradient using the cycling parameters in Table 1. The PCR products were separated on a 2% agarose gel and a 250bp ladder was used as DNA molecular weight standard.14,15
The vacA gene detectionThe presence of the genotypes of vacA alleles (s1, s2, m1 and m2) was determined by multiplex PCR using the primers in Table 1. DNA samples from J99 H. pylori strain was used as positive controls and sterile distilled water was used as a negative control. The 20μL reaction mixture consisted of 1× PCR buffer, 1.5mM magnesium chloride, 400μM dNTP, 0.5pmol of each primer, 1U hot start taq polymerase (Promega) and 4μL of template DNA. Amplification was carried out in an Eppendorf Mastercycler gradient using the cycling parameters in Table 1. The PCR products were separated on a 2% agarose gel and a 100bp ladder was used as DNA molecular weight standard.14
Statistical analysisThe data were analyzed using SPSS software (Version 17.SPSS Inc, United States) and p values were calculated using Chi-square test to find any significant relationship. p<0.05 was considered statistically significant.
ResultsIsolates and antibiotic susceptibility testingOf two hundred patients, H. pylori infection was found in 163 individuals (81.5%) on the basis of positive rapid urease test. A total of 151 (76%) H. pylori were isolated and the antibiotic susceptibility was determined. According to EUCAST breakpoints, only 26 isolates were susceptible to all antibiotics and all isolates tested were susceptible to tetracyclin (MIC range, 0.016–0.064mg/L), rifampicin (MIC range, 0.016–0.032mg/L) and amoxicillin (MIC range, 0.016–0.064mg/L. The resistance rate to clarithromycin (MIC range, 0.5–32mg/L) was (25.2%, n=38) and for ciprofloxacin (MIC range, 1–32mg/L), it was (18.5%, n=28). The isolates were characterized by high rates of resistance to metronidazole (MIC range, 8–256mg/L) (67.5%, n=102) (Table 2). Twenty six isolates were resistant to both metronidazole and clarithromycin. Twelve of the 28 ciprofloxacin-resistant isolates were also resistant to metronidazole and one was resistant to clarithromycin. Metronidazole-resistant isolates were found more frequently in women compared with men (43.05% versus 24.4%, p<0.05) but there were no relation between the gender or age and resistance for the other antibiotics (p>0.5).
Resistance to antibiotics of H. pylori isolates in males and females from 2012 to 2015, Algeria.
| Antimicrobial | Resistance (%) (n/total) | Resistance in females (%) (n/total) | Resistance in males (%) (n/total) |
|---|---|---|---|
| AMX | 0 (0/151) | 0 (0/151) | 0 (0/151) |
| CIP | 18.5 (28/151) | 9.93 (15/151) | 8.61 (13/151) |
| CLA | 25.2 (38/151) | 15.89 (24/151) | 9.27 (14/151) |
| MTZ | 67.5 (102/151) | 43.05 (65/151) | 24.50 (37/151) |
| RIF | 0 (0/151) | 0 (0/151) | 0 (0/151) |
| TET | 0 (0/151) | 0 (0/151) | 0 (0/151) |
AMX, amoxicillin; CLA, clarithromycin; CIP, ciprofloxacin; MTZ, metronidazole; RIF, rifampicin; TET, tetracycline.
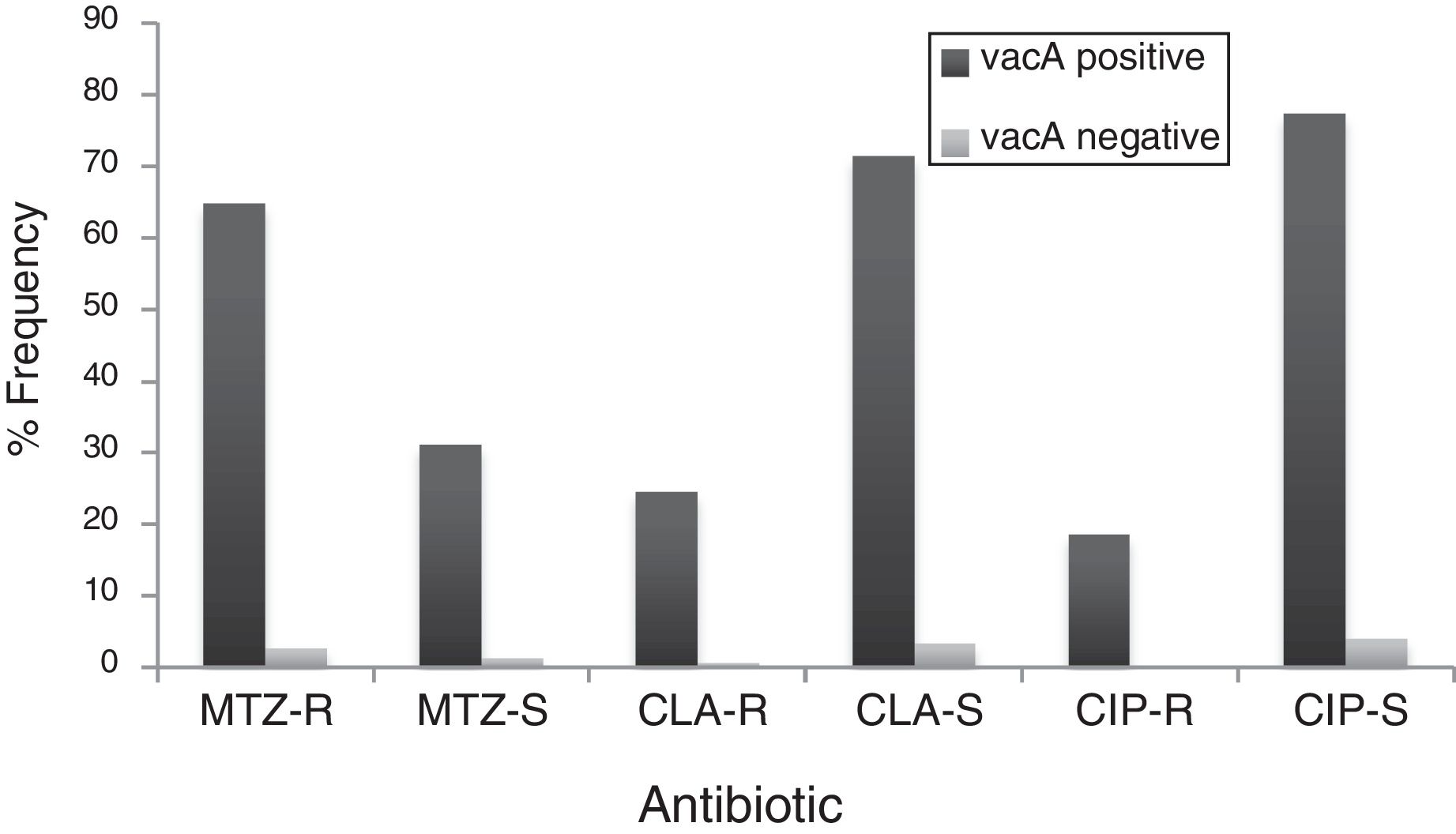
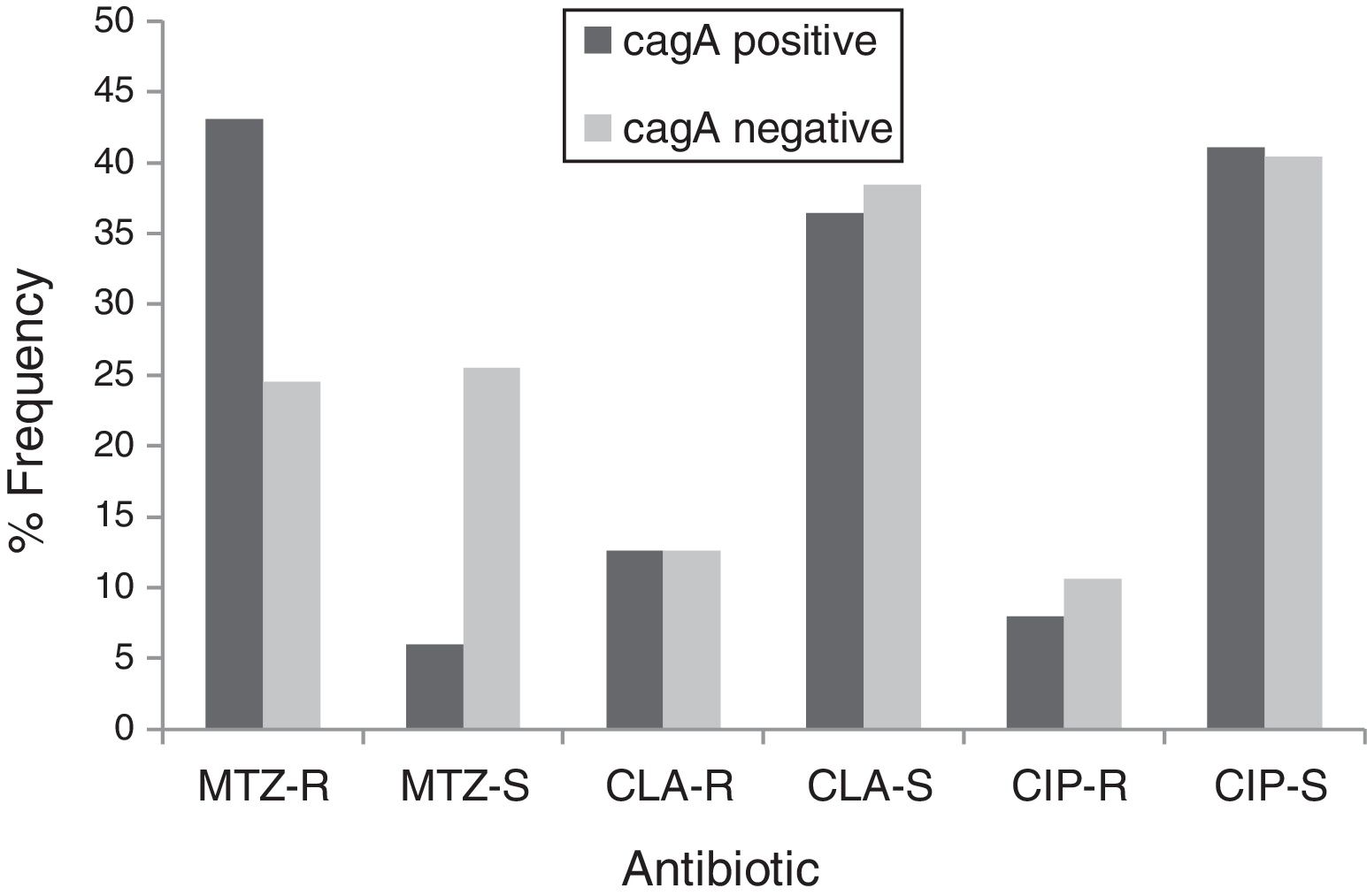
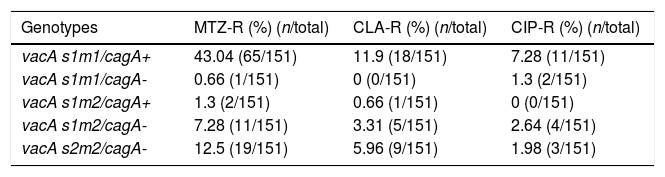
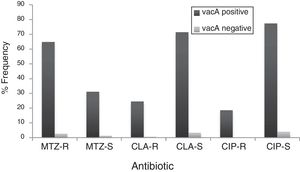
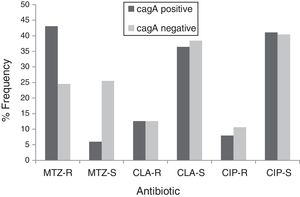
By PCR H. pylori was detected in 167 out of 200 (83.5%) where both 16S rRNA and glmM genes were detected. Generally, of 167 positive gastric biopsy samples, 97 samples (58%) were cagA positive and all samples had amplified bands for both vacA s and m regions. Overall, 100 (59.88%) samples had vacA s1m1, 30 (17.96%) had vacA s1m2, 37 (22.15%) had vacA s2m2. No statically significant relationship was found between vacA or cagA genotypes and antibiotic resistance results (p>0.5) except for the metronidazole, where it had relation with the presence of cagA genotype (p=0.001) (Figs. 1 and 2). Among the resistant isolates, the vacA s1m1/cagA+ genotype was the most prevalent followed by the vacA s1m1/cagA− genotype (Table 3).
Prevalence of antibiotic-resistant in vacA positive and vacA negative H. pylori isolates. MTZ-R, metronidazol resistant; MTZ-S, metronizaol susceptible; CLA-R, clarithromycin resistant; CLA-S, clarithromycin susceptible; CIP-R, ciprofloxacin resistant; CIP-S, ciprofloxacin susceptible.
Prevalence of antibiotic-resistant in cagA positive and cagA negative H. pylori isolates. MTZ-R, metronidazol resistant; MTZ-S, metronizaol susceptible; CLA-R, clarithromycin resistant; CLA-S, clarithromycin susceptible; CIP-R, ciprofloxacin resistant; CIP-S, ciprofloxacin susceptible.
Resistance to antibiotics of H. pylori genotypes.
| Genotypes | MTZ-R (%) (n/total) | CLA-R (%) (n/total) | CIP-R (%) (n/total) |
|---|---|---|---|
| vacA s1m1/cagA+ | 43.04 (65/151) | 11.9 (18/151) | 7.28 (11/151) |
| vacA s1m1/cagA- | 0.66 (1/151) | 0 (0/151) | 1.3 (2/151) |
| vacA s1m2/cagA+ | 1.3 (2/151) | 0.66 (1/151) | 0 (0/151) |
| vacA s1m2/cagA- | 7.28 (11/151) | 3.31 (5/151) | 2.64 (4/151) |
| vacA s2m2/cagA- | 12.5 (19/151) | 5.96 (9/151) | 1.98 (3/151) |
H. pylori infection represents the most prevalent chronic bacterial disease in the world with a range that affects more than half of the population.16 The prevalence of H. pylori differs significantly between countries, with high rates of infection being associated with low socioeconomic status and high densities of living. For instance, in Japan, South America, Turkey and Pakistan, the prevalence is more than 80%, while in England it is lower (20%).17H. pylori was detected in 167 (83.5%) of a total of 200 biopsy specimens by the PCR method. Our findings are similar to other reports in Iran.18 This rate is higher than the rate found when using culture (76%). This is due to the sensitivity and the specificity of the PCR and the exigency of the bacterium in culture conditions which makes its isolation very difficult without forgetting that culture has the great advantage of allowing performance of antimicrobial susceptibility.
The glmM gene is highly conserved and has been used to identify H. pylori in gastric biopsies. It has better sensitivity than the ureA gene.19 One of the advantages of using the glmM gene to identify H. pylori directly in gastric biopsies is its high degree of sensitivity and specificity, because it has a detection rate of 10–100 H. pylori cells, which is better than histopathology and culture.20 Our findings revealed a rate of positive H. pylori in the tested biopsies of 83.5% based on direct molecular detection by PCR using the glmM genes. Although it is referred that glmM gene detection is more sensitive than other genes, in this study, detection of 16S rRNA gene had the same sensitivity as glmM gene. Hoshina et al. also used 16S rRNA sequences for the specific detection of H. pylori. The specificity of H. pylori detection reported by Hoshina and colleagues is similar to our results.21
In Algeria, the prevalence of H. pylori is very high so it is very important to know the rates of its resistance to antibiotics, especially when antibiotic resistance represents a serious public health problem in Algeria and around the world. In adults, the rates of primary resistance to clarithromycin and metronidazole were documented as 15% for clarithromycin and 37% for metronidazole in 2013.3 There is an increase in the rate of clarithromycin resistance with 25.2% and the same for metronidazol (67.5%) found in our present study. Concerning clarithromycin primary resistance, our prevalence would seem higher than that found in Congo (1.7%),22 in Iceland (9%)23 and Tunisia (14.6).24 This rate was almost similar to those found in China (28.9%),25 and in Spain (17.1%).26 This difference in clarithromycin resistance rate between several countries might be due to the prescription of this antibiotic. Since clarithromycin is widely used as antimicrobial drug to heal infections in other organ systems such as respiratory system and previous consumption of macrolides (cross resistance), the prevalence of clarithromycin-resistant is continuously increasing.27
Metronidazole resistance is higher in the developing countries (50–80%). Our result was much higher than that found in Europe (33.1%)28,29 and in Iceland (1%),23 but was similar to the rate found in China (63.8%)25 and lower than that found in Columbia (82%).30 We found a higher primary metronidazole resistance in female patients compared with male ones, most likely due to the wide use of this antibiotic for gynecological infections. In accordance with data presented by Glupczynski et al. and Batnavala et al., metronidazole resistant isolates were found altogether more frequently in females than in males and in non European natives.31,32 Koletzko et al. also found that metronidazole was widely prescribed for infections such as parasitic or female genital infections in Africa and in Asia which explains our results.33
Fluoroquinolone resistance was found in 28 out of 151 (18.5%). In Asia, different values among countries were detected, the resistance rate being 14.9% in Japan,34 and 28% in China.25 In a single Italian study,35 levofloxacin resistance was higher with old (>45 years) than young patients (28.4% vs. 14.4%). In Korea the resistance rate of H. pylori to ciprofloxacin was reported to be (33.8%),36 this data agree with our results. But in Sweden a low rate of resistance (3.3%) was reported with ciprofloxacin.37 This increasing resistance rate could be explained by the use of fluoroquinolones for urinary infections and a cross resistance between the different molecules of this antibiotic group.
No amoxicillin, rifampicin and tetracyclin resistance was observed. For amoxicillin resistance finding, it was similar to those determined by Wolle et al.38 in their study in Japan. In Europe, available data from two studies enrolling 599 patients found a prevalence rate <1%.28 A high prevalence was reported In Asia and South America (38%).39,40 Amoxicillin is widely used in Algeria but the absence of resistance could be due to the complexity of amoxicillin resistance mechanisms (pbp gene mutations, membrane permeability alterations, efflux pumps, etc.). Rifampicin resistance stains are rare; this was confirmed in two studies, showing the presence of a resistant strain in 22 out of 1585 patients in Germany (1.4%),41 and in 17 out of 255 patients in England (6.6%; 95%).42 This antibiotic is rarely used for H. pylori eradication treatment which explains the absence of resistant strains in Algeria. In accordance with our study, tetracycline resistance was absent in patients from Iceland,23 and very low in Congo (2.5%) and in China (3.9%).22,25 In contrast, increased values were found in Chilly (27%).43 Unfortunately, tetracycline-based regimens require the use of bismuth salts which are no longer available in several countries due to possible side-effects.
It is evident now that the presence of H. pylori virulence genes and their different genotypic combinations in the strain colonizing a patient, affects the development of the gastric disease. Some characteristics of H. pylori strains have been associated with the progression of infection to more severe disease. Some genotypes, such as alleles s1 and m1 of the vacA gene and presence of the cagA gene, are considered pathogenicity markers since they are associated with cytotoxin production and the induction of more intense epithelial lesions and inflammatory reactions. The cagA was identified in 58% patients, this result is higher to those obtained in Pakistan (24.2%),44 but lower than those obtained in Tunisia of 61.6%,45 Iran (76%) and Iraq (71%).46 The cagA positive strains in a Mexican study were 86%.47 In Japan, the rate of cagA is very high (90%),48 which is correlated with the high prevalence of gastric cancer in that country.
The majority of H. pylori isolates in our study were vacA positive. The virulence of the isolate has strong relation with the mosaic combination of s and m region allelic types which is responsible of the production of the cytotoxin; the predominant combination of vacA alleles in our study was s1/m1 (59.88%). These results are in accordance with several other studies. In Brazil,49 and in Mixico,50vacA s1/m1 was the predominant genotype in these countries too. Numerous studies have provided evidence for a stronger association of disease outcomes in individuals infected with H. pylori strains possessing active vacA genotypes such as s1 and m1 than that with less or non-active vacA types.51,52
Concerning cagA and vacA alleles combination, we found that the predominant was cagA+/s1m1. This finding is in accordance with other studies performed in Latin America such us Faundez et al. who found that cagA+/s1m1 strains were the most common in Brazilian patients53 and Morales-Espinosa et al. also found that cagA+/s1m1 strains had a higher prevalence in Mexican patients.54 Additionally, this genotype is the most prevalent in Asian population too.51cagA is most commonly associated with the vacA s1 genotype, particularly in patients with peptic ulcer or gastric cancer. Therefore, the cagA+ vacA s1 genotype is present in patients with high expression of tumor necrosis factor receptor-associated factor 1, tumor necrosis factor receptor superfamily member 9, or B-cell lymphoma-extra-large.55 In our study, we have selected patients with upper abdominal pain, this explain our low rate of 58% cagA and 59.88% s1/m1 which is low comparably with other countries, the prevalence of these genotypes in patients with peptic ulcer and gastric cancer is significantly greater than among those with gastritis alone.53
The relation of virulence factors and the resistance results describes no direct role of vacA or cagA genes in antibiotic resistance which agrees with Ghotaslou et al. results. The vacA s1m1/cagA+ genotype is associated with more severe gastric diseases due to the fact that strains with this genotype cause severe inflammation with increased production of IL-1β and TNF-α, inducing inhibitors of secretion of hydrochloric acid and increase gastric pH; conditions that may favor the action of antibiotics.56 The metronidazole resistance had relation with the presence of cagA genotype in our study, this data disagree with those reported by Khan et al.57 where they found that the absence of cagA gene contributes in the acquisition of resistance. Apparently, infected patients with vacA s1m1/cagA+ resistant strains are at increased risk of progression to more severe conditions by failure in H. pylori eradication. Thus, in this population, infected patients with vacA s1m1/cagA+ resistant strains are at increased risk of severe disease than infected patients with vacA s1m1/cagA− or vacA s2m2/cagA−, however, all must be closely monitored.
In patients from Algeria with upper abdominal pain, the prevalence of H. pylori is 83.5%, 67.5% of the isolates were resistant to metronidazole, 25.2% were resistant to clarithromycin and 18.5% were resistant to ciprofloxacin. These rates of resistance to metronidazole and clarithromycin are higher than internationally accepted ranges and allow stopping triple therapy for H. pylori eradication and new therapeutic strategies are needed in Algeria. Metronidazole, might well be refrained from using in order to prevent the emergence of more resistant strains especially because of the high rate of metronidazole resistance in Algeria. About clarithromycin, this antibiotic should not be used, or a clarithromycin susceptibility test should be performed, before prescription. Therefore, it is necessary to monitor the evolution of fluroquinolone resistance in this area. The vacA s1m1/cagA+ genotype was the most prevalent among resistant isolates. There was no significant difference found in resistance with H. pylori genotypes. It is likely that infected patients with resistant strains vacA s1m1/cagA+ are at increased risk of progression to more severe conditions by failure in H. pylori eradication. These data are of a great importance for the regional health systems of Algeria.
EthicsThis work is registered in a national research project entitled: University research project (CNEPRU), Financed by the Algerian Research Ministry and has this following registration code: D01N01UN020120150001. This study was approved by the ethics committees of all hospitals where the endoscopy was performed and all patients provided written informed consent before endoscopy.
FundingThis work was financed by “Laboratoire de bioressources”, Department of Biology, Faculty of Sciences, Hassiba Ben Bouali University, 02000 Chlef, Algeria.
In Algeria, all research projects are financed by the Algerian Research Ministry through research laboratories in each University. Project grant number D01N01UN020120150001.
Authors’ contributionsConceived and designed the experiments: MB, RA, AT and MM. Performed the experiments: MB, AT, AED and MM. Analyzed the data: MB, AT and MD. Contributed reagents/materials/analysis tools: MB, MM and KTD. Wrote the paper: MB, RA. All authors read and approved the final manuscript.
Conflicts of interestThe authors declare no conflicts of interest.
We thank Dr Samir Halouche (Clinique de Gastroenterology, 02000 Chlef, Algeia) and Pr Bousria (First November 1954 University Hospitol, Oran, Algeria) for their technical assistance, and also the clinicians involved in this study. We thank Dr Hacene Mahmoudi and Dr Ahmad Aichouni for their moral support.