This study was carried out to express human epidermal growth factor (hEGF) in Pichia pastoris GS115. For this aim, the hEGF gene was cloned into the pPIC9K expression vector, and then integrated into P. pastoris by electroporation. ELISA-based assay showed that the amount of hEGF secreted into the medium can be affected by the fermentation conditions especially by culture medium, pH and temperature. The best medium for the optimal hEGF production was BMMY buffered at a pH range of 6.0 and 7.0. The highest amount of hEGF with an average yield of 2.27μg/mL was obtained through an induction of the culture with 0.5% (v/v) methanol for 60h. The artificial neural network (ANN) analysis revealed that changes in both pH and temperature significantly affected the hEGF production with the pH change had slightly higher impact on hEGF production than variations in the temperature.
Human epidermal growth factor (hEGF) is a 6.2kDa polypeptide composed of 53 amino acid residues with three intramolecular disulfide bonds.1 One of its major biological functions is to promote the generation of new epithelial and endothelial cells, and to stimulate tissue repairs.2 As such, it has a powerful mitogenic activity, which can speed up the healing process of damaged tissues due to, among other things, ulcers and wounds. hEGF was also found to be effective in the treatments of wrinkles, age spots, freckles and acnes.3,4 Traditionally, hEGF is purified from animal urine through a complicated purification step.5 However, the yield and purity of the hEGF from this process are low and could not meet the demand from the industries. Genetic engineering is one approach in which pure hEGF can be potentially produced in a large scale. Over the last two decades, hEGF had been produced in various host systems including Escherichia coli,6Bacillus brevis,7Saccahromyces cerevisiae8 and baculovirus.9 In E. coli, the yield is not up to the level appropriate for the industrial needs, as cytoplasmic hEGF tends to form inclusion bodies, which can be rapidly degraded by proteases, leading to the formation of misfolded hEGF molecules. As a result, the overall production cost is increased due to the extra downstream processing steps required to release the hEGF from the inclusion bodies. In addition, the hEGF produced by prokaryotic system exhibits lower biological activities compared to those produced by eukaryotic systems. Therefore, eukaryotic expression systems may hold a promising host for the production of active hEGF if post-translational processing is really needed especially in a large-scale production.
Among the eukaryotic expression systems, Pichia may offer a cheaper alternative host for hEGF production. Pichia pastoris is widely applied as host systems for the expressions of many heterologous proteins. As a methylotrophic yeast, P. pastoris is able to grow in the presence of methanol; therefore, methanol can be used an inducer without any toxic effects. In the present study, the secretary expression vector (pPIC9K) equipped with a strong methanol inducible promoter (AOX1) was used to express and secrete hEGF extracellularly.10,11 Pichia has both the advantages of E. coli and eukaryotic expression systems for being an inexpensive host system, able to highly express heterologous proteins, and able to modify proteins post-transnationally.
Materials and methodsMicroorganisms and plasmidsE. coli DH5α, as a host for construction of DNA plasmids, was cultured in Luria Bertani (LB) medium supplemented with 50μg/mL ampicillin. P. pastoris strain GS115, as a host for the production of hEGF, cultured in YPD medium [1% (w/v) yeast extract, 2% (w/v) peptone, 2% (w/v) dextrose (glucose)] at 30°C. DNA plasmid pFLAG-ATS-hEGF, obtained from the Faculty of Biotechnology and Biomolecular Sciences, UPM, Malaysia, was used a template for the amplification of hEGF gene. The pPIC9K expression plasmid (Invirtrogen, USA) was used for the expression of hEGF in P. pastoris.
Construction of pPIC9K-hEGF DNA plasmidThe hEGF gene (GeneBank Accession No. M15672) was amplified from the previously constructed plasmid pFLAG-ATS-hEGF12 using the specific primers 5′-GAATTCATGAACTCAGATAG-3′ and 5′-CCTAGGTCAACGTAATTCC-3′. Amplification was carried out in a thermocycler (Gene Amp PCR system, BioRad, USA); after denaturation at 95°C for 2min, the sample was subjected to 35 cycle of 95°C for 30s, 57°C for 30s, and 72°C for 20s followed by final extension at 72°C for 5min. The amplified fragment was then inserted into the EcoRI and AvrII cloning sites of the yeast expressing plasmid pPIC9K. After transformation of the recombinant plasmids into E. coli DH5α cells, gene insertion was checked by restriction enzyme digestion and PCR. The integrity of the insert was checked by double-stranded sequencing (1st BASE DNA Sequencing Services, Malaysia).
Expression of hEGF in P. pastorisElectro-transformation of pPIC9K-hEGF into P. pastorisA single colony of P. pastoris GS115 was cultured in 5mL YPD medium in a 50mL conical flask overnight in a shaker incubator at 30°C with 250rpm agitation rate. The culture was transferred into fresh YPD medium (250mL) and incubated under the same condition until the OD600 nm reached 1.3–1.5. The cells were then harvested at 1500×g for 5min at 4°C; following 2 times washing with 500 and 250mL ice-cold sterile dH2O, the cells were resuspended in 20mL ice-cold 1M sorbitol. Finally, the cells were centrifuged and resuspended in 1mL ice-cold 1M sorbitol. For electro-transformation, in a 0.2cm cuvette (BioRad, USA), the P. pastoris competent cells were mixed with 0.5μg of the recombinant pPIC9K-hEGF plasmid; after 5min incubation on ice, electroporation was carried out at 1.5kV. Immediately after electroporation, 1mL of ice-cold 1M sorbitol was added to the cells; in a sterile microtube, the cells were mixed with 1mL YPD medium and incubated as at 30°C for 1h. Around 200μL of the mixture was spread on MD agar plates [1.34% yeast nitrogen base with ammonium sulfate without amino acids (YNB), 4×10−5% biotin and 2% dextrose]. The plates were incubated at 30°C until recombinant colonies were appeared (2–5 days).
Direct screening of multiple insertsPositive transformants were screened on the YPD agar supplemented with geneticin at the final concentrations of 0.25, 0.5, 0.75, 1.0, 1.5, 1.75, 2.0, 3.0 and 4mg/mL. Positive clones were screened by direct colony PCR as previously described by Linder et al.13 with some modifications. Briefly, a single colony of the transformants was picked and resuspended in 10μL of sterile dH2O. Then, 5μL of 5U/μL solution of lyticase was added to the cells; after 10min incubation at −80°C, the extract was subjected to PCR using the 5′ and 3′ AOX1 primers according to the following cycles: 95°C for 5min for pre-denaturation, 30 cycles of amplification at 95°C for 1min, 54°C for 1min (annealing) and 72°C for 1min followed by a final extension step at 72°C for 5min.
In vivo screening of multiple insertionsHis+ clones were resuspended in dH2O. Then, the suspensions were transferred to 50mL Falcon tube, vortexed for 8s and cell density was determined at 600nm using a spectrophotometer. The cells were placed on YPD containing 0.25, 0.5, 0.75, 1.0, 1.5, 1.75, 2.0, 3.0 and 4mg/mL geneticin, separately. The plates were incubated at 30°C and monitored for survivals every day.
Production of recombinant hEGFIn a 250mL baffled flask, a single colony of the recombinant P. pastoris was inoculated in 25mL BMGY [buffered glycerol-complex medium: 1% (w/v) yeast extract, 2% (w/v) peptone, 100mM potassium phosphate, pH 6.0, 1.34% (w/v) yeast nitrogen base (YNB), 4×10−5% (w/v) biotin] containing 1% glycerol (v/v) followed by overnight incubation at 30°C in a shaking incubator (250rpm) until the optical density at 600nm reached 2 (log-phase growth). The cells were then harvested at 2500×g for 5min at RT; the pellet was resuspended in 150mL of BMMY medium supplemented with 0.5% methanol and incubated until the optical density at 600nm reached 1.0. In a 1L baffled flask, the culture was further incubated at 29°C with agitation at 270rpm for 96h. In order to maintain the induction, methanol was added to a final concentration of 0.5% every 24h. The culture was checked for hEGF production at 0, 6, 12, 24, 36, 48, 60, 72, 84 and 96h of incubation.
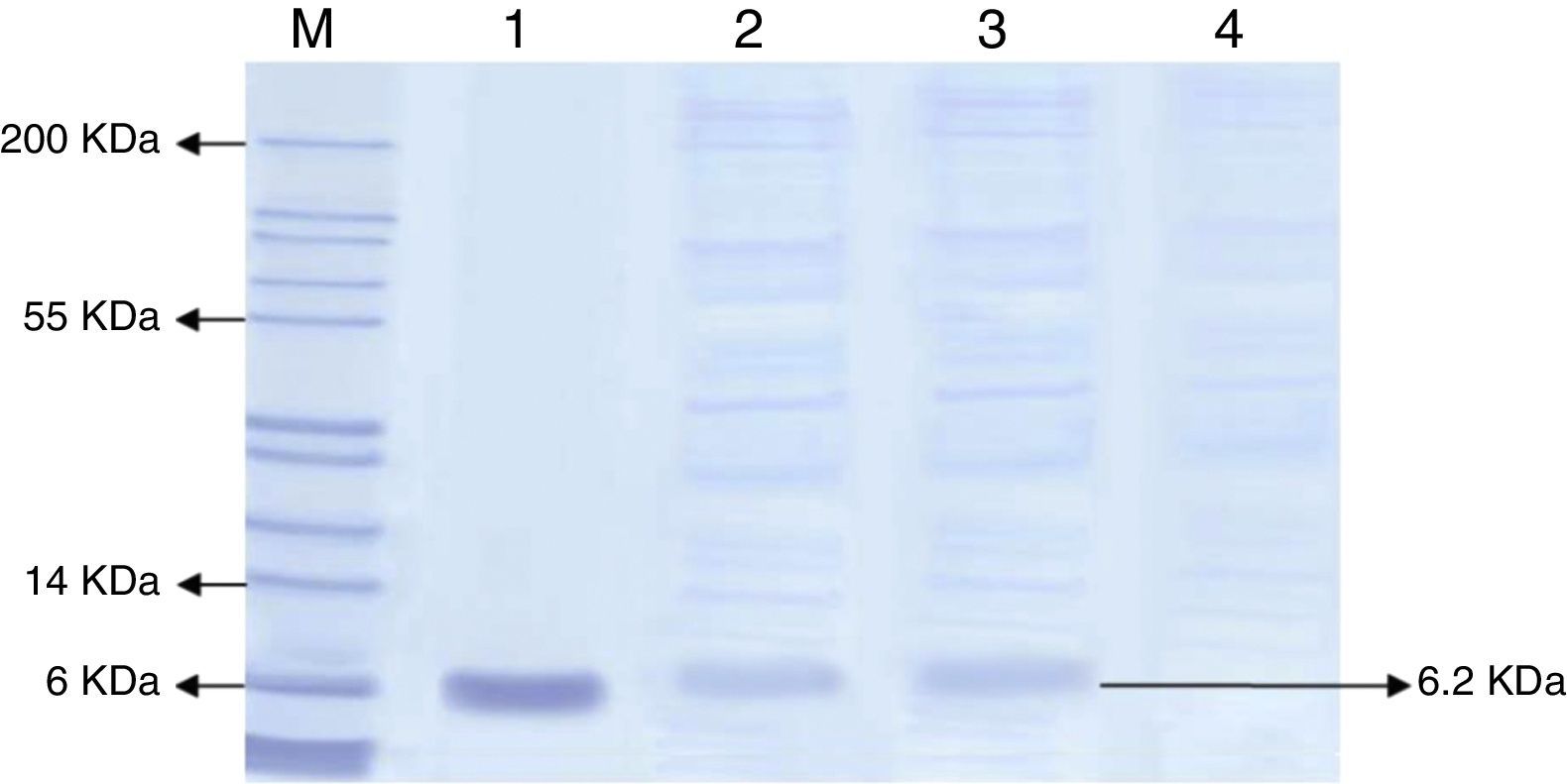
SDS-PAGE analysisSecreted proteins were separated on a 12% SDS-polyacrylamide gel. Eighty micro-liter of each sample was boiled in a 20μL sample buffer (312mM Tris–HCL pH 6.8, glycerol 50% (v/v), bromophenol blue 0.05% (w/v) and dH2) for 10min before the sample mixture were being loaded onto the gel. Standard hEGF protein (65.0kDa) (R&D SYSTEM) was used as a positive control. hEGF protein was purified directly from the SDS gel using a commercially available polyacrylamide gel protein purification kit (Thermo Scientific).
Quantikine hEGF immunoassayFor hEGF expression assay, the Quantikine Human EGF immunoassay kit (R&D system, USA) was used according to the manufacture's instruction. A standard curve was created by plotting the mean absorbance at 570nm for each standard on the y-axis against the concentration on the x-axis. The amount of hEGF was determined by referring the absorbance value of the samples at 570nm to the hEGF standard curve. The data were linearized by plotting the EGF concentrations versus the log of the absorbance and the best-fit line was determined by regression analysis.
Effect of culture conditions on growth and hEGF productionCulture mediumIn 1-L baffled flask, the cells were grown on 150mL of YPD, BMMY, MM or BMGY. Cell biomass and hEGF production were determined at 0, 12, 24, 36, 48, 60, 72 and 84h after culturing. The growth condition used in this experiment was similar with the one described earlier. Methanol was used at a final concentration of 0.5% (MM and BMMY) every 24h in order to maintain a continual induction and its final concentration. The best medium was selected for further optimization as below.
Methanol concentrationThe recombinants were grown in the same condition as previously described. Different concentrations of methanol [0, 0.25, 0.5, 1, 1.5 and 2% (v/v)] were evaluated. The respective final concentrations of methanol were maintained by adding the appropriate amounts of methanol to the cultures for every 24h. Samplings of the cultures were done every 12h as described above.
pHOptimization was done in two steps; in the first step, a large range of pH from 4 to 7 was used; in the second step, the pH range was further fine-tuned based on the two best pH obtained from the first step. The cultures were incubated using the same culture condition that was described before. The samples were taken every 12h for analysis.
Combination of pH and temperatureThe range of pH used in this study was between 4 and 7 comprising four different levels of pHs i.e. 4.0, 5.0, 6 and 7; whereas the effect of temperature was evaluated at 25°C, 30°C, 32°C and 35°C. A total of six combinations were performed and the growth condition used was the same with the one mentioned before. The artificial neural network (ANN) was used to optimize pH and temperature for hEFG production. We used ANN with three layers: (1) an input layer with 2 neurons, (2) a hidden layer with 7 neurons, and (3) an output layer with 1 neuron. The topology of the network contained three layers (2:7:1): one input layer for two fermentation variables, pH and temperature; middle hidden layer of 7 neurons, and one output layer for hEGF production.
Statistical analysisThe data were analyzed by using one way analysis of variance followed by liner correlation tests where a p value of less than 0.05 was considered as significant. The statistical analysis of the ANN data and plots were performed using the Statistical software version 7.
ResultsConstruction of pPIC9K-hEGF expression plasmidThe hEGF gene was amplified from the pFLAG-ATS-hEGF plasmid and inserted into the expression vector pPIC9K. After transformation into E. coli DH5α, positive clones were selected with direct PCR screening and restriction enzyme digestion. Finally, the identity of the inserts were checked by double-stranded sequencing pPIC9K-hEGF plasmid was sequenced and compared to the original hEGF sequence. The comparison analysis showed the cloned hEGF was 100% similar to the original hEGF (GeneBank Accession No. M15672).
Generation of recombinant P. pastorisThe DNA plasmid construct pPIC9K-hEGF was linearized by SacI and electro-transformed into P. pastoris. The recombinant clones were screened on YPD supplemented with different concentrations of geneticin from 0.25 to 4mg/mL. According to the results (data not shown), most of the transformants were found to be resistant to the low concentration of Geneticin (0.25, 0.5 and 0.75mg/mL) and just a few were resistant to the high concentrations. Direct PCR proved successful insertion of the hEGF gene into P. pastoris genome (Data not shown).
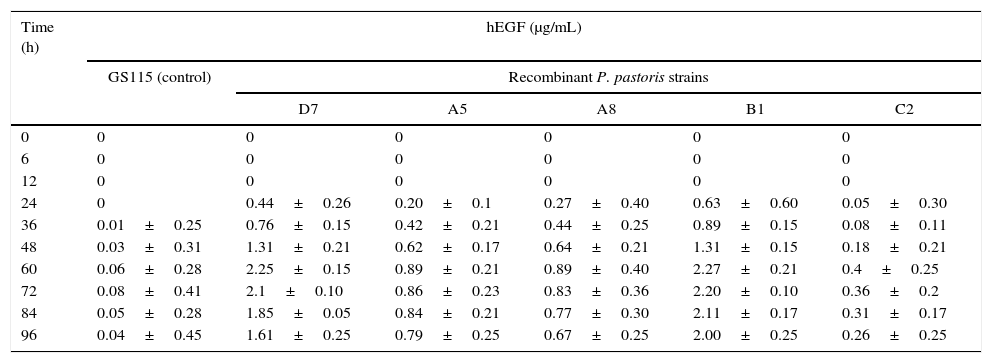
Production of recombinant hEGFFive recombinant clones selected from the hyper-resistant clones were tested for the production of hEGF by ELISA at ten different time points after induction. As shown in Table 1, the highest level of hEGF production was obtained 60h post-induction. Very weak absorbance readings considered as background reading of the ELISA assay were observed in the control sample (GS115). Both B1 and D7 clones showed the highest hEGF production, 60h post-induction, however, in D7, the level of hEGF was found to be decreased rapidly. Therefore, the B1 clone was selected for the optimization of fermentation conditions and medium compositions.
hEGF production in different clones of P. pastoris.
| Time (h) | hEGF (μg/mL) | |||||
|---|---|---|---|---|---|---|
| GS115 (control) | Recombinant P. pastoris strains | |||||
| D7 | A5 | A8 | B1 | C2 | ||
| 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 6 | 0 | 0 | 0 | 0 | 0 | 0 |
| 12 | 0 | 0 | 0 | 0 | 0 | 0 |
| 24 | 0 | 0.44±0.26 | 0.20±0.1 | 0.27±0.40 | 0.63±0.60 | 0.05±0.30 |
| 36 | 0.01±0.25 | 0.76±0.15 | 0.42±0.21 | 0.44±0.25 | 0.89±0.15 | 0.08±0.11 |
| 48 | 0.03±0.31 | 1.31±0.21 | 0.62±0.17 | 0.64±0.21 | 1.31±0.15 | 0.18±0.21 |
| 60 | 0.06±0.28 | 2.25±0.15 | 0.89±0.21 | 0.89±0.40 | 2.27±0.21 | 0.4±0.25 |
| 72 | 0.08±0.41 | 2.1±0.10 | 0.86±0.23 | 0.83±0.36 | 2.20±0.10 | 0.36±0.2 |
| 84 | 0.05±0.28 | 1.85±0.05 | 0.84±0.21 | 0.77±0.30 | 2.11±0.17 | 0.31±0.17 |
| 96 | 0.04±0.45 | 1.61±0.25 | 0.79±0.25 | 0.67±0.25 | 2.00±0.25 | 0.26±0.25 |
Based on the highest level of hEGF protein production, the presence of hEGF in the culture filtrate was further analyzed by SDS-PAGE, as shown in Fig. 1. The hEGF protein band represented the expected size of the protein (6.2kDa) as compared to the standard hEGF and the marker. The total amount of secreted proteins in the medium was also determined by the Bradford assay, where 10.4mg/mL protein was detected for the B1 strain. The hEGF protein was then purified.
SDS-PAGE of total protein secreted from the wild and recombinant strain of P. pastoris. The SDS gel was stained by Coomassine blue. M: stained protein ladder; lane 1: standard hEGF protein; lanes 2 and 3: samples of recombinant P. pastoris B1 clone; lane 4: sample of wild type P. pastoris GS115. Each well was loaded with 100μL of culture filtrate from recombinant P. pastoris B1 and wild type P. pastoris.
The growth curve of the recombinant P. pastoris showed a short lag phase, lasting for a few hours, followed by the exponential growth for 50h, before reaching the stationary phase 10h later (Fig. 2). The first appearance of hEGF was observed after 24h of culture i.e. shortly after being induced at 20h. A rapid hEGF production occurred concomitantly with an increase in the growth. Under this condition, the hEGF was continually produced even after the culture reached its stationary phase.
Optimization of culture condition for hEGF productionBaffled flask was used for optimization of the culture condition for growth and hEFG production in P. pastoris.
Culture mediumA time course investigation of hEGF production and biomass accumulation in different media in shake flasks was carried out where the highest hEGF production was obtained in BMMY and MM medium, as shown in Fig. 3. In term of biomass production, all of the media were found to support good growth of the culture. Although, the best cell biomass production was observed in the YPD medium, this medium failed to promote good production of hEGF.
Methanol concentrationThe effect of different concentrations of methanol (0.0%, 0.25%, 0.5%, 1.0%, 1.5% and 2%) on growth of the recombinant P. pastoris and hEGF production are presented in Fig. 4A and B. A decrease in the production of hEGF and cell biomass was detected, when the concentration of methanol was either too low or too high, although the growth rate was found to be higher only at relatively low concentration (0.25% of the final concentration). The highest level of hEGF production was observed when the concentration of methanol was 0.5% (v/v).
A time-course hEGF production and cell biomass of recombinant P. pastoris after being induced by different concentrations of methanol. (A) Effects of different methanol concentrations on hEGF production. (B) Effects of methanol concentrations on growth of the recombinant yeast. Data are means of biological triplicates. 0% methanol (
), 0.25% methanol (), 0.5% methanol (), 1% methanol (), 1.5% methanol () and 2% methanol ().As shown in Fig. 5, higher yield of hEGF was detected at a pH range of 6–7. Even though Pichia can grow on a pH range of 4 and 5, which is slightly acidic, the amount of hEFG was low. Fig. 6 shows the effect of a narrow pH range on hEGF production. The result also showed that the recombinants could grow in a wide pH range from 3.0 to 7.0, however the growth rate was found to be lower at pH below 6.0.
Data of the combination effects of pH and temperature analyzed by ANN, as shown in Table 2, revealed significant effects of both pH and temperature on hEGF production. The optimum pH and temperature calculated by the ANN were found to be 7 and 29°C, respectively. The predicted amount of hEGF production at these levels was 2.371μg/mL. The combinatorial effects of pH and temperature were represented by the surface plot where changes in the culture media pH showed more impact on the hEGF production than temperature (Fig. 7A). The analysis of ANN sensitivity was carried out to analyze the ratio and ranking of the variables where variables with a ratio higher than one were considered as significant.14 Based on this, both parameters significantly influenced the production of hEGF (Table 3). The effect of individual factor on hEGF production is shown in Fig. 7B and 7C. The production of hEGF was increased as the pH increased higher than 4.0. The optimal temperature for hEGF production was 29°C and the levels of hEGF was expected to be decreased when the temperature was higher than 31°C.
Effect of different pH and temperature on hEGF production.
| Time (h) | hEGF (μg/mL) | |||||
|---|---|---|---|---|---|---|
| 25°C | 30°C | |||||
| pH | ||||||
| 4 | 5 | 6.5 | 4 | 5 | 6.5 | |
| 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 12 | 0 | 0 | 0 | 0 | 0 | 0 |
| 24 | 0 | 0 | 0.45 | 0.09 | 0.25 | 0.27 |
| 36 | 0 | 0.21 | 0.78 | 0.11 | 0.45 | 1.05 |
| 48 | 0.07 | 0.25 | 1.35 | 0.22 | 0.49 | 1.62 |
| 60 | 0.11 | 0.40 | 2.21 | 0.26 | 0.65 | 2.48 |
| 72 | 0 | 0.19 | 2.17 | 0.18 | 0.44 | 2.44 |
| 84 | 0 | 0.05 | 2.10 | 0.12 | 0.30 | 2.37 |
This study was carried out to produce recombinant hEGF in P. pastoris GS115 using a commercially available expression vector equipped with a specific signal peptide that directs the transportation of the hEGF protein into the culture medium. The ability of P. pastoris to use methanol as a sole carbon and energy source allows the use of methanol alcohol oxidase gene (AOX1) promoter for the expression of the desire gene.15,16 Glycerol was used as a sole carbon source until they reached the log-phase growth where methanol was supplemented in the medium for hEGF induction as suggested.17
The hEGF gene was inserted into the pPIC9K expression vector, and then integrated into the yeast by electroporation. The clones were screened on YPD supplemented with different concentrations of geneticin from 0.25 to 4mg/mL. The level of geneticin resistance depends on the copy number of the kanamycine gene integrated into the Pichia genome and it seems that multiple integrations of the recombinant gene in the Pichia genome increase the expression of the desired protein.18–21 Therefore, clones resistant to the high concentration of geneticin were selected for the production of recombinant hEGF.
ELISA-based assay confirmed successful secretion of hEGF in the BMMY medium. Highest level of hEGF was detected in the hyper-resistant clone B1, 60h post-induction.
The amount of hEGF secreted into the medium was shown to be affected by the fermentation condition. Optimization of culture conditions is one of the determining factors to the success of recombinant protein production in yeast strains. Shaken flask, small-scale expression methods are often the first step in order to optimize the protein expression levels and selecting the best culture conditions,22 although the level of protein production from shake flask cultures is 10-fold lower than the level that would be achieved using a fermenter because of its lower cell density23 and limited aeration.24,25
Highest level of hEGF was detected in BMMY and MM medium. It can be due to the presence of methanol in these media which acts a sole source of carbon and energy as well as an inducer for the gene expression system. It is a common practice for high production of recombinant protein, high densities culture is used to promote high yield protein production based on the fact that the level of recombinant protein production is proportional to cell density.11 However, this was not happened in this study, as YPD medium which promoted good growth of the yeast culture failed to induce hEGF production. BMMY which support good growth and highest level of hEGF production was selected as the cultivating medium for the recombinant B1 strain. This medium has been shown to be also the best medium for the expression of SAG2 and bikunin.26,27
Optimization of methanol concentration in Pichia fermentation system is extremely important because a high level of methanol can be toxic to the yeast.28 In some cases, increasing methanol concentration causes increasing in the expression level such as expression of human β-2-glycoprotein I.29 However the accumulation of methanol has a negative result of cell growth and can lead to a decrease in the expression.25,30 On the other hand, a low level of methanol may not be enough to initiate the transcription of the recombinant gene.10 Methanol concentration of 0.5% (v/v) was considered as the optimal concentration for the production of hEGF which is similar to what has been reported for laccase production in P. pastoris host system.31 It was shown that methanol with concentration higher than 0.5% (v/v) might inhibit growth whereas very low concentration of methanol might not be enough to support energy metabolism leading to poor growth of cultures. In fact, methanol was not only used to provide a sole carbon source for Pichia but also to induce hEGF production. However, the levels of methanol should be kept low to avoid the toxicity effect of methanol,32 which would inhibit cell growth and protein production. The toxicity effect of methanol was evident at 1% (v/v) with the poor growth rate of the culture, which in turn reduced the hEGF production.
Like temperature, pH of culture media plays an important role for optimized heterologous protein production as it is not only affect the yield of heterologous protein production but also affect the integration of the protein structure.33 In addition, the pH of culture media would also affect the activities of various proteases that may degrade heterologous proteins. Our results showed that the recombinant Pichia could grow in a wide pH range from 3.0 to 7.0, although highest yield of hEGF was obtained at the pH range of 6–7. This feature is an extra advantage of the yeast for recombinant protein production where a wide range of pH can be used for the optimization of the recombinant protein production as demonstrated in this study. Data analysis by ANN showed the significant effects of both pH and temperature on hEGF production, although changes in pH were found to be more effective. ANN analysis recommended 29°C as the optimal temperature for hEGF production in P. pastoris and the yield of hEGF was expected to be decrease at temperatures higher than 31°C, possibly due to poor stability of the protein at higher temperatures, releasing of proteases from dead cells, and misfolding of the protein.34,35
In conclusion, this study supports the use of P. pastoris as a host for hEGF production, specifically when the overall hEGF production is compared with the production in E. coli. The BMMY medium with pH 7 supplemented with 0.5% (v/v) methanol is recommended as the best induction medium for hEGF production in P. pastoris.
Conflicts of interestThe authors declare no conflicts of interest.
This work was supported by the faculty of Biotechnology and Biomolecular Sciences, Universiti Putra Malaysia, Malaysia.