Plant growth-promoting rhizobacteria strains from special formulations have been used to optimize eucalyptus cutting production. To undertake quality control for the formulated products, the rhizobacterial strains should be characterized to assess their purity and authentication. In the present study, we characterized nine strains of rhizobacteria, including three Bacillus subtilis (S1, S2 and 3918), two Pseudomonas sp. (MF4 and FL2), P. putida (MF2), P. fulva (Ca), Frateuria aurantia (R1), and Stenotrophomonas maltophilia (CIIb). The strains were differentiated by colony morphology after 24h of incubation in three different solid state culture media (glucose-nutritive agar, 523 medium and yeast extract-mannitol agar), sensitivity to a panel of 28 antibiotics (expressed according to the formation of inhibition halos of bacterial growth in the presence of antibiotics), and PCR-RFLP profiles of the 16S rDNA gene produced using nine restriction enzymes. It was possible to differentiate all nine strains of rhizobacteria using their morphological characteristics and sensitivity to antibiotics. The molecular analysis allowed us to separate the strains CIIb, FL2 and R1 from the strains belonging to the genera Bacillus and Pseudomonas. By using these three methods concomitantly, we were able to determine strain purity and perform the authentication.
Free-living bacteria or bacteria associated with root tissues prevail in the plant rhizosphere.1Plant growth-promoting rhizobacteria (PGPR), the benefic group of these microorganisms, are a class of non-pathogenic soil microorganisms.2–4 Rhizobacteria are natural inhabitants of soil that are able to colonize the root systems of plants, thereby contributing several important characteristics. For example, enhanced growth can occur directly through the production of growth promoters, or it can be inhibited by the action of pathogenic microorganisms.5
PGPR bacteria may directly influence plant growth by either synthesizing plant hormones, such as indol-3-acetic acid (IAA),6,7 or favoring the uptake of nutrients from the soil through different mechanisms, such as nitrogen fixation,8 phosphorus and potassium solubilization9 and the synthesis of siderophores for iron sequestration.10 PGPR can also indirectly affect plants through antagonism between the bacteria and soil-borne pathogens11 and by inducing systemic resistance in plants against both root and foliar pathogens.
Many studies have explored the biocontrol capacity of these organisms. Additionally, their ability to produce antibiotics makes them a target for the biological control of plant diseases. Strains of rhizobacteria isolated from Eucalyptus spp. have been shown to promote rooting through an increase in root biomass and growth of eucalyptus cuttings12,13 and the reduction of Cylindrocladium cutting rot, rust infection (Puccinia psidii Winter) in the nursery,14,15 and bacterial wilt (Ralstonia solanacearum).16 Based on the results, a bioproduct named Rizolyptus®17 formulated with selected rhizobacterial strains has been used in eucalyptus cutting nurseries. The Rizolyptus® is an inoculant based on only one rhizobacteria strain in a liquid formulation. However, it is essential to know the intrinsic characteristics of each selected growth-promoting rhizobacteria strain prior to mass propagation to ensure the product's quality.
Bacterial characterization is currently based on biochemical tests, antibiotic sensitivity, microscopic observations and molecular analysis. Restriction fragment length polymorphism (RFLP) analysis of the ribosomal DNA region (rDNA) associated with polymerase chain reaction (PCR) is an appropriate and inexpensive molecular method.1,2,18–20 Thus, in the present work, we characterized the Rizolyptus® production from nine rhizobacterial strains based on their morphology, antibiotic sensitivity, and PCR-RFLP profiles. The findings and methods presented in this study represent important tools to ensure the purity, quality and authentication of strains in the final product Rizolyptus® for commercialization.
Material and methodsRhizobacterium strainsNine strains of rhizobacteria isolated from eucalyptus that were previously selected for their capacity to promote rooting and the growth of eucalyptus cuttings14 were characterized. The strains were identified and coded as follows: S1, S2 and 3918 (Bacillus subtilis Cobn); Ca (Pseudomonas fulva Lizuga & Komagata); CIIb (Stenotrophomonas maltophilia Hugh, Palleroni & Bradbury); R1 (Frateuria aurantia Swings et al.); MF2 (P. putida Migula); and FL2 and MF4 (Pseudomonas sp. Migula). The molecular identification, based on homology (>98%) of the 16S rDNA was performed as previously described.21 The cultures of rhizobacteria are stored in the Forest Pathology Laboratory/Bioagro of the Universidade Federal de Viçosa, Minas Gerais, Brazil.
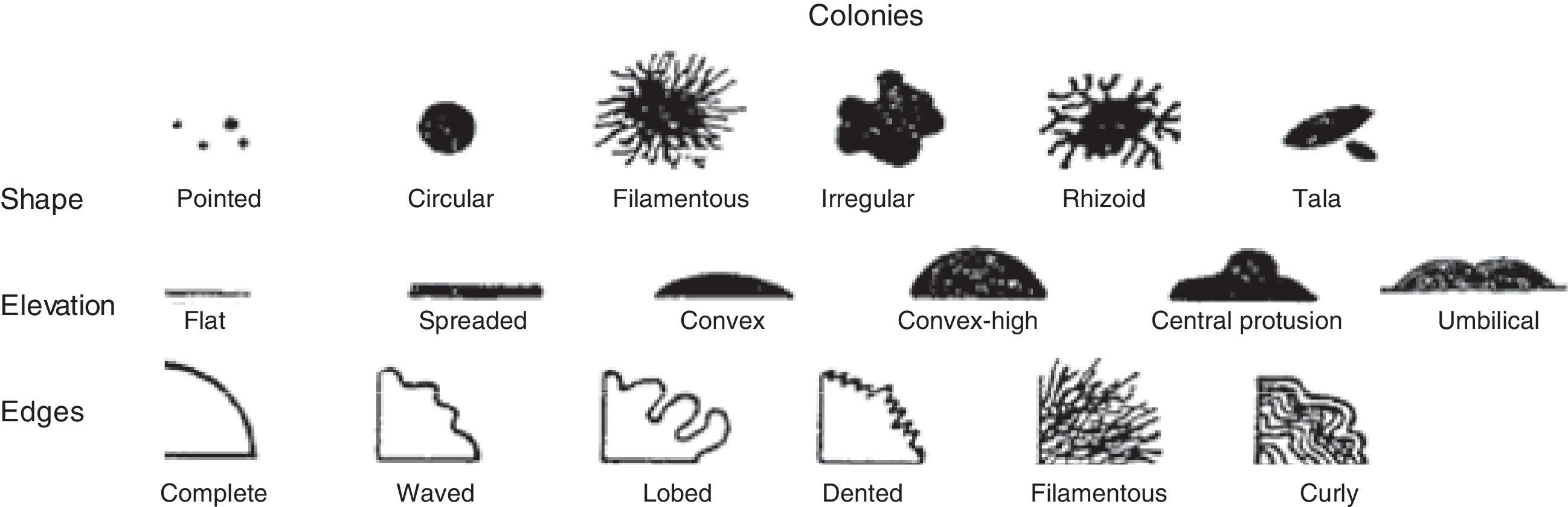
Morphological characterizationThe strains were grown on 523 medium,22 yeast extract-mannitol agar (YMA)23 and glucose-nutritive agar (ANG)24 for 24h at 28°C and were characterized according to their colony shape, elevation, edge type (Fig. 1), consistency (i.e., mucosus, fluid or mycelial), aspect of the colony surface (i.e., smooth or rough), brightness (i.e., bright, translucent or opaque), color, size (i.e., <1mm, 1–2mm, 2–3mm or >3mm), and growth speed (i.e., very fast: visible to the naked eye after less than 24h of incubation; fast: visible within 24–48h; intermediate: visible within 24–48h; slow: visible within 36–96h; or very slow: visible only after 96h).
Patterns used for the morphological characterization of rhizobacterial strains based on their colony shape, elevation and edge type.
Strain sensitivity to 28 antibiotics was assessed using the standard antibiogram method.25 An inoculum sample of 0.1mL was evenly spread in a Petri dish (9cm diameter) containing 523 medium, and four Whatman® No.1 filter paper disks (Ø=0.7cm) that were previously soaked in the antibiotics to be tested were distributed over the medium. A completely random design containing three replicates per antibiotic was used. After a 48-h incubation, the presence or absence of an inhibition halo was observed.
Molecular characterizationGenomic DNA from the rhizobacterial strains26 was amplified by a PCR reaction consisting of 10–20ng DNA, 2mM MgCl2, 50mM KCl, 10mM Tris–HCl (pH 8.3), 0.1mM of each deoxynucleotide (dATP, dTTP, dCTP and dGTP) (Invitrogen), 0.1μM of each oligonucleotide, 1 unit of Taq polymerase (Phoneutria) enzyme and sterile water (MilliQ) to reach the final volume of 50μL. The specific oligonucleotides P1 (5′-AGA GTT TGA TCC TGG CTC AG-3′) and P2 (5′-AAG GAG GTG ATC CAG CCG CA-3′)27 were used to amplify a fragment of approximately 1.6kb from the bacterial 16S rDNA 16S region. The amplification was performed in a Mastercycler® thermocycler (Eppendorf) under the following conditions: 94°C for 3min, then 35 cycles comprising 1min at 94°C, 30s at 60°C and 1min and 30s at 72°C, and a final extension at 72°C for 7min. Subsequently, 15μL of the amplified product was cleaved separately with the EcoRI, RsaI, Sau3AI, SphI, MspI, BamHI, DdeI, TaqI and HinfI restriction enzymes. A volume of 1μL of restriction enzyme in 3μL of 10× buffer was added to the amplified product and incubated under the specific time and temperature conditions recommended for each enzyme by the manufacturer (Promega). The cleaved fragments were separated in an agarose gel (1.2%) and photodocumented.
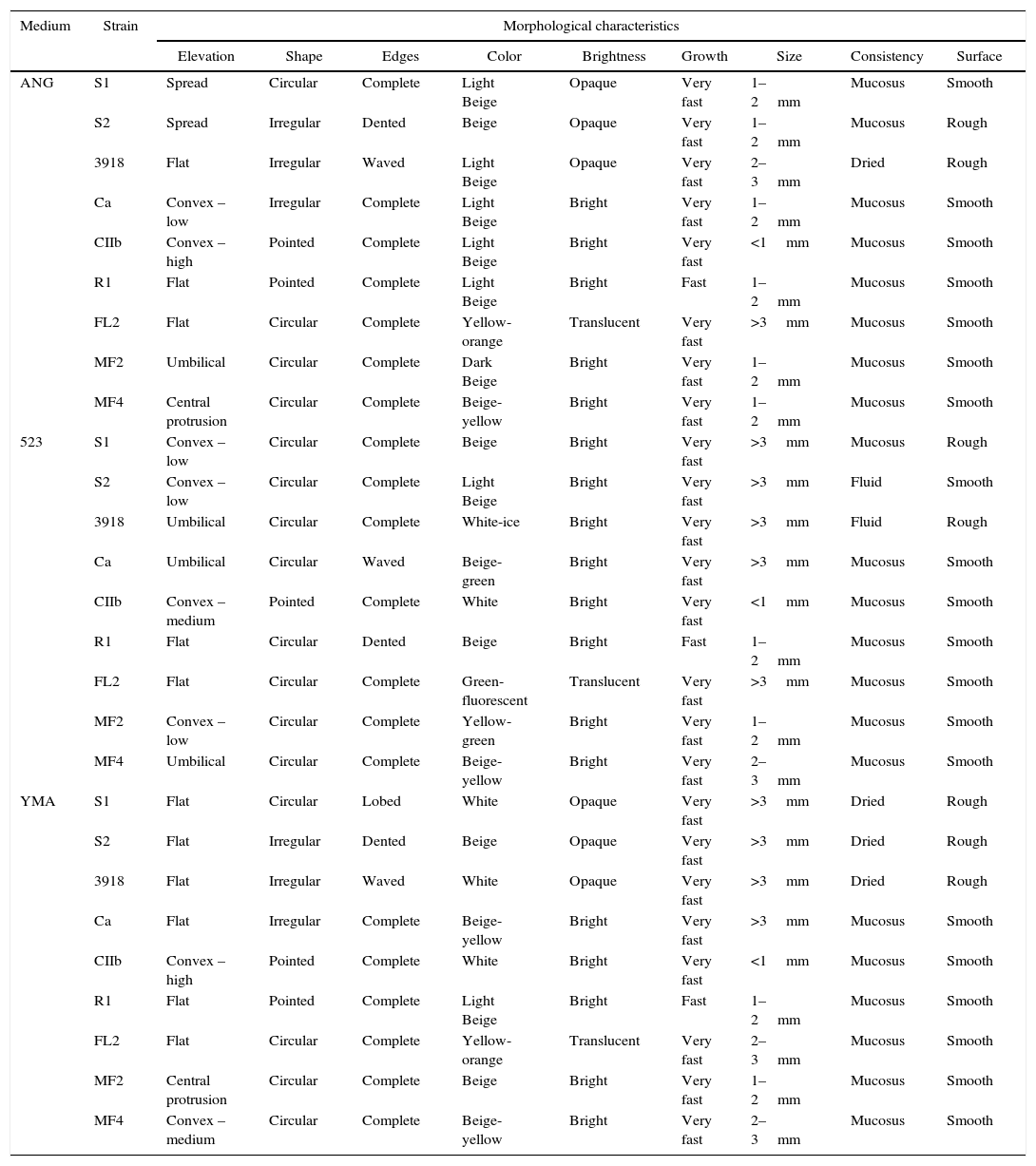
ResultsMorphological characterizationThe morphological characterization varied according to the culture medium (523, YMA, or ANG) and the rhizobacterial strain assessed. The colony color in 523 medium allowed for the differentiation of eight of the nine strains tested, whereas the colony elevation in ANG medium allowed for the differentiation of six strains (Table 1). All of the strains could be differentiated on 523 medium when these two characteristics were combined. Consistency, surface, and growth speed provided minor contributions to the differentiation of the assessed rhizobacterial strains (Table 1).
Morphological characterization of rhizobacterial strains assessed after 24h of incubation in solid state glucose-nutritive agar (ANG), 523 medium and yeast extract-mannitol agar (YMA).
| Medium | Strain | Morphological characteristics | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Elevation | Shape | Edges | Color | Brightness | Growth | Size | Consistency | Surface | ||
| ANG | S1 | Spread | Circular | Complete | Light Beige | Opaque | Very fast | 1–2mm | Mucosus | Smooth |
| S2 | Spread | Irregular | Dented | Beige | Opaque | Very fast | 1–2mm | Mucosus | Rough | |
| 3918 | Flat | Irregular | Waved | Light Beige | Opaque | Very fast | 2–3mm | Dried | Rough | |
| Ca | Convex – low | Irregular | Complete | Light Beige | Bright | Very fast | 1–2mm | Mucosus | Smooth | |
| CIIb | Convex – high | Pointed | Complete | Light Beige | Bright | Very fast | <1mm | Mucosus | Smooth | |
| R1 | Flat | Pointed | Complete | Light Beige | Bright | Fast | 1–2mm | Mucosus | Smooth | |
| FL2 | Flat | Circular | Complete | Yellow-orange | Translucent | Very fast | >3mm | Mucosus | Smooth | |
| MF2 | Umbilical | Circular | Complete | Dark Beige | Bright | Very fast | 1–2mm | Mucosus | Smooth | |
| MF4 | Central protrusion | Circular | Complete | Beige-yellow | Bright | Very fast | 1–2mm | Mucosus | Smooth | |
| 523 | S1 | Convex – low | Circular | Complete | Beige | Bright | Very fast | >3mm | Mucosus | Rough |
| S2 | Convex – low | Circular | Complete | Light Beige | Bright | Very fast | >3mm | Fluid | Smooth | |
| 3918 | Umbilical | Circular | Complete | White-ice | Bright | Very fast | >3mm | Fluid | Rough | |
| Ca | Umbilical | Circular | Waved | Beige-green | Bright | Very fast | >3mm | Mucosus | Smooth | |
| CIIb | Convex – medium | Pointed | Complete | White | Bright | Very fast | <1mm | Mucosus | Smooth | |
| R1 | Flat | Circular | Dented | Beige | Bright | Fast | 1–2mm | Mucosus | Smooth | |
| FL2 | Flat | Circular | Complete | Green-fluorescent | Translucent | Very fast | >3mm | Mucosus | Smooth | |
| MF2 | Convex – low | Circular | Complete | Yellow-green | Bright | Very fast | 1–2mm | Mucosus | Smooth | |
| MF4 | Umbilical | Circular | Complete | Beige-yellow | Bright | Very fast | 2–3mm | Mucosus | Smooth | |
| YMA | S1 | Flat | Circular | Lobed | White | Opaque | Very fast | >3mm | Dried | Rough |
| S2 | Flat | Irregular | Dented | Beige | Opaque | Very fast | >3mm | Dried | Rough | |
| 3918 | Flat | Irregular | Waved | White | Opaque | Very fast | >3mm | Dried | Rough | |
| Ca | Flat | Irregular | Complete | Beige-yellow | Bright | Very fast | >3mm | Mucosus | Smooth | |
| CIIb | Convex – high | Pointed | Complete | White | Bright | Very fast | <1mm | Mucosus | Smooth | |
| R1 | Flat | Pointed | Complete | Light Beige | Bright | Fast | 1–2mm | Mucosus | Smooth | |
| FL2 | Flat | Circular | Complete | Yellow-orange | Translucent | Very fast | 2–3mm | Mucosus | Smooth | |
| MF2 | Central protrusion | Circular | Complete | Beige | Bright | Very fast | 1–2mm | Mucosus | Smooth | |
| MF4 | Convex – medium | Circular | Complete | Beige-yellow | Bright | Very fast | 2–3mm | Mucosus | Smooth | |
The R1 strain was differentiated from the others by its fast growth (within 24h) following incubation in all three growth media (Table 1). Colonies of the CIIb isolate differed from the others based on their pointed shape and colony size of less than 1mm in the three media tested. Although the S1, S2 and 3918 strains belong to B. subtilis, they were individually differentiated by their color in 523 medium and by their edge shape in ANG and YMA media (Table 1). These strains were separated from the others by their brightness in ANG and YMA media and by their surface characteristics in YMA medium. The Ca, FL2, MF2 and MF4 strains were differentiated from the others by colony color. Additionally, FL2 was the only strain that showed a translucent brightness in the three culture media evaluated (Table 1).
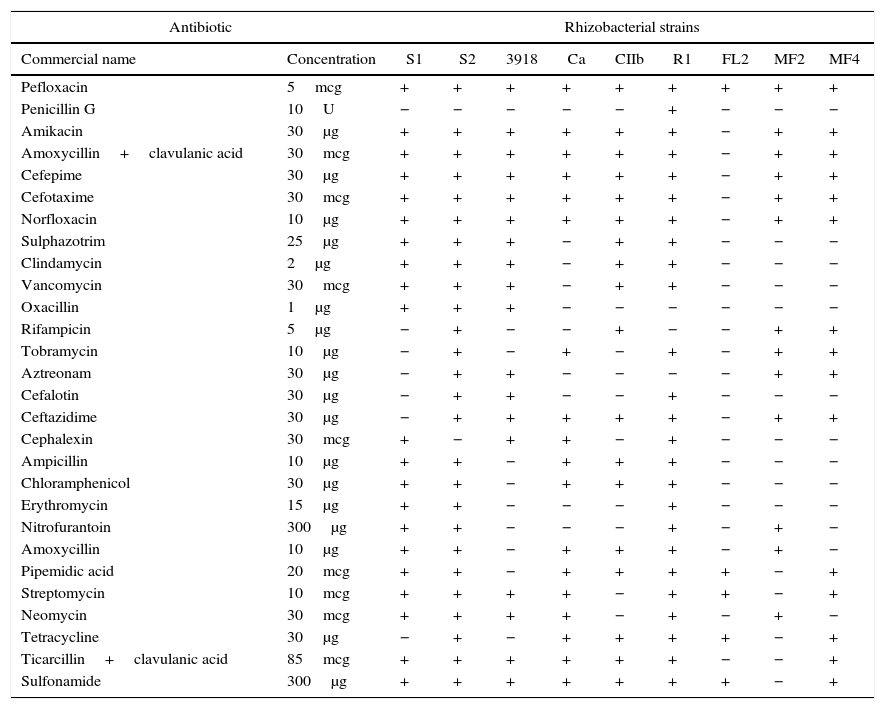
Antibiotic sensitivityPefloxacin was the only antibiotic tested that inhibited all of the rhizobacterial strains (Table 2). All of the remaining antibiotics inhibited at least one strain. The S2 and R1 strains were the most sensitive to the antibiotics tested and were resistant to only two (penicillin and cephalexin) and three (oxacillin, rifampicin, and aztreonam) antibiotics, respectively. In contrast, FL2 was the least sensitive, with only five antibiotics (pefloxacin, pipemidic acid, streptomycin, tetracycline, and sulfonamide) inhibiting its growth. The MF2 and FL2 strains were easily separated from each other and the other strains using a single antibiotic (e.g., sulfonamide and amikacin, respectively). A minimum of two antibiotics were required to distinguish between the S1, CIIb, and MF4 strains (e.g., for the 1st strain: ceftazidime and tetracycline; 2nd strain: streptomycin and neomycin; and 3rd strain: amoxycillin and rifampicin). The other strains could be distinguished using at least three antibiotics sequentially in the culture medium (e.g., for S2: cephalexin, sulphazotrim, and streptomycin; 3918: ampicillin, sulfonamide, and neomycin; and Ca: clindamycin, sulfonamide, and amikacin).
Rhizobacterial strain sensitivity expressed according to the formation of inhibition halos of bacterial growth in the presence of antibiotics.
| Antibiotic | Rhizobacterial strains | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Commercial name | Concentration | S1 | S2 | 3918 | Ca | CIIb | R1 | FL2 | MF2 | MF4 |
| Pefloxacin | 5mcg | + | + | + | + | + | + | + | + | + |
| Penicillin G | 10U | − | − | − | − | − | + | − | − | − |
| Amikacin | 30μg | + | + | + | + | + | + | − | + | + |
| Amoxycillin+clavulanic acid | 30mcg | + | + | + | + | + | + | − | + | + |
| Cefepime | 30μg | + | + | + | + | + | + | − | + | + |
| Cefotaxime | 30mcg | + | + | + | + | + | + | − | + | + |
| Norfloxacin | 10μg | + | + | + | + | + | + | − | + | + |
| Sulphazotrim | 25μg | + | + | + | − | + | + | − | − | − |
| Clindamycin | 2μg | + | + | + | − | + | + | − | − | − |
| Vancomycin | 30mcg | + | + | + | − | + | + | − | − | − |
| Oxacillin | 1μg | + | + | + | − | − | − | − | − | − |
| Rifampicin | 5μg | − | + | − | − | + | − | − | + | + |
| Tobramycin | 10μg | − | + | − | + | − | + | − | + | + |
| Aztreonam | 30μg | − | + | + | − | − | − | − | + | + |
| Cefalotin | 30μg | − | + | + | − | − | + | − | − | − |
| Ceftazidime | 30μg | − | + | + | + | + | + | − | + | + |
| Cephalexin | 30mcg | + | − | + | + | − | + | − | − | − |
| Ampicillin | 10μg | + | + | − | + | + | + | − | − | − |
| Chloramphenicol | 30μg | + | + | − | + | + | + | − | − | − |
| Erythromycin | 15μg | + | + | − | − | − | + | − | − | − |
| Nitrofurantoin | 300μg | + | + | − | − | − | + | − | + | − |
| Amoxycillin | 10μg | + | + | − | + | + | + | − | + | − |
| Pipemidic acid | 20mcg | + | + | − | + | + | + | + | − | + |
| Streptomycin | 10mcg | + | + | + | + | − | + | + | − | + |
| Neomycin | 30mcg | + | + | + | + | − | + | − | + | − |
| Tetracycline | 30μg | − | + | − | + | + | + | + | − | + |
| Ticarcillin+clavulanic acid | 85mcg | + | + | + | + | + | + | − | − | + |
| Sulfonamide | 300μg | + | + | + | + | + | + | + | − | + |
Absence of inhibition halo (−).
Presence of inhibition halo (+).
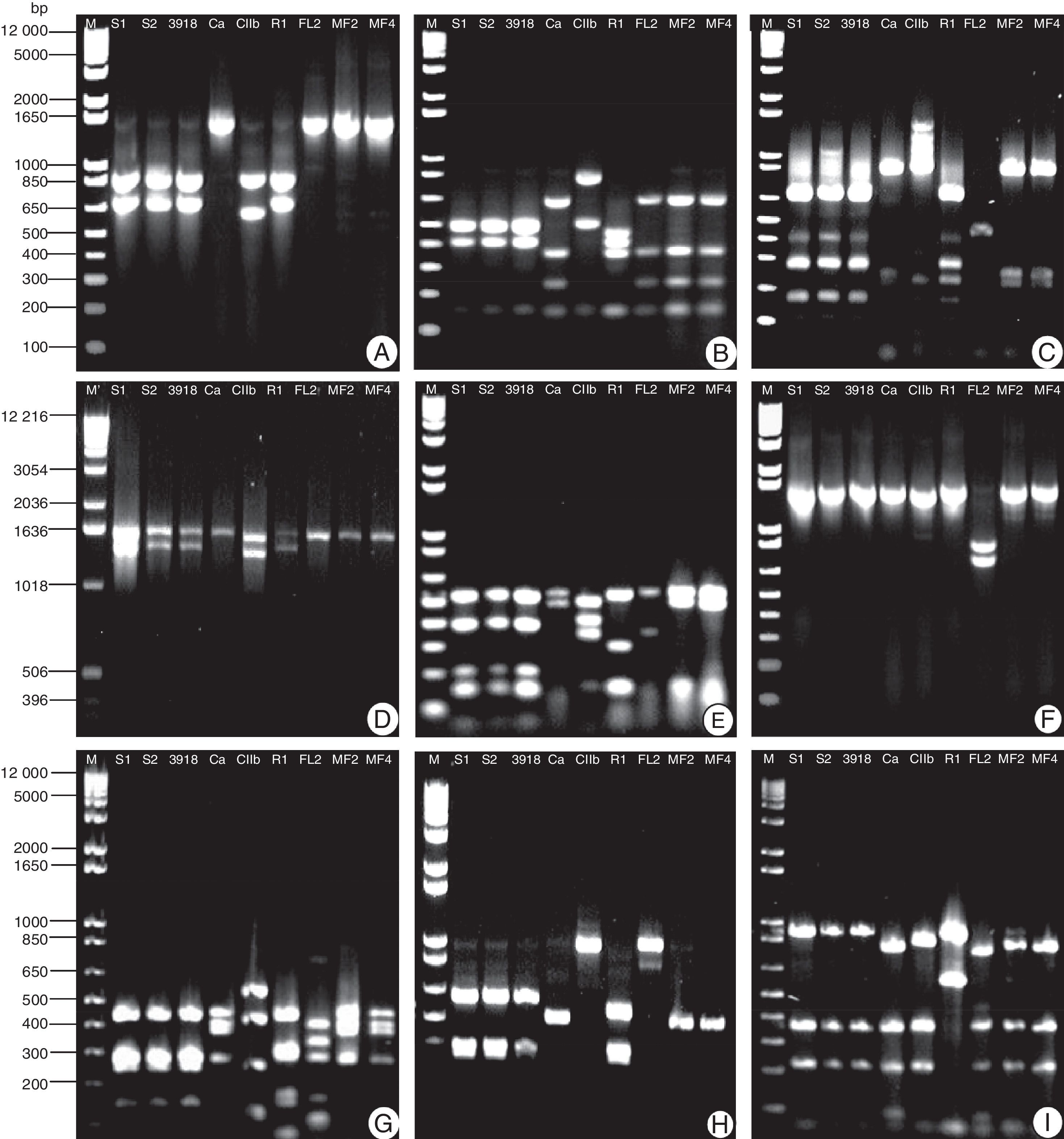
The amplification of 16S rDNA generated fragments approximately 1.6kb in size for all strains. The cleavage for these fragments with restriction enzymes allowed us differentiate two groups of strains (Bacillus (3918, S1, and S2) and Pseudomonas (MF2, MF4, and Ca)), from the remaining strains (Fig. 2). However, it was not possible to distinguish strains from the same genus. All of the other strains could be differentiated from each other using at least one restriction enzyme. CIIb was the most unique among the tested strains, with six of the nine tested enzymes resulting in restriction profiles that allowed for its separation from the other strains. Although Ca and FL2 belong to the same genus, they were easily differentiated by cleavage with the Sau3AI, MspI, DdeI, BamHI, and TaqI enzymes. These rhizobacteria belong to different species of Pseudomonas, which makes their identification easier; similar results were observed for Stenotrophomonas maltophilia (CIIb) and Frateuria aurantia (R1). The MspI enzyme allowed us to discern more strains, separating Bacillus (3918, S1, and S2), Pseudomonas (MF2, MF4, and Ca), FL2, R1, and CIIb. In contrast, the use of BamHI only differentiated FL2 from the other strains. The remaining enzymes allowed us to separate the strains into 3–5 groups (Fig. 2).
Restriction profile of amplified fragments of the 16S region from the rDNA of nine rhizobacterial strains. The fragments were cleaved with the following restriction enzymes: (A) EcoRI, (B) RsaI, (C) Sau3AI, (D) SphI, (E) MspI, (F) BamHI, (G) DdeI, (H) HinfI, (I) TaqI, and (M) restriction enzyme – 1kb Plus DNA Ladder.
The use of pre-selected bacterial strains to induce rooting and growth of eucalyptus cutting requires authentication of the rhizobacteria employed in the formulated product (Rhizolyptus®) without mixture between the strains of rhizobacteria or contamination. In this study, we characterized nine strains of rhizobacteria based on their morphology, antibiotic sensitivity, and PCR-RFLP profiles.
All strains were distinguished based on their morphological characteristics in culture and sensitivity to 28 antibiotics. Their morphological characteristics varied according to the culture medium and strain used. Color and colony elevation in 523 medium were the most efficient morphological features for strain identification, allowing the differentiation of all rhizobacterial strains studied. However, the use of these characteristics demanded expertise and skill to distinguish subtle differences among the strain colonies. Special care was also required to recognize intermediate shapes that may hamper the accuracy of the identification. Therefore, adding antibiotics to the culture medium could aid in the identification of bacterial strains. For example, the antibiotic sensitivity profile and triage by PCR of the toxin showed that Bacillus pumilus was the predominant species in the seashore area around Cochin, India.28 In addition to help the rhizobacterial identification, the supplementation of the culture medium used to produce the inoculants with antibiotics avoided contamination or strain misuse because it allowed only antibiotic-resistant strain growth. For example, the P. putida (MF2) strain could be differentiated by the use of sulfonamide to ensure strain purity because this antibiotic inhibited all of the other rhizobacterial strains tested. Similarly, amikacin, amoxycillin and clavulanic acid, cefepime, cefotaxime, and norfloxacin did not affect the growth of the FL2 isolate.
Recently, molecular techniques have been broadly employed in taxonomy and biodiversity studies of bacteria.1,19,20,29–32 The PCR-RFLP analysis results may vary among genera and species along with the 16S rRNA and 23S rRNA sequences.1,32 We used the 16S gene in the PCR-RFLP technique for the molecular characterization of rhizobacterial strains because it yields consistent results and is an easy to use, reliable, and sensitive method. The results from the molecular analysis in this study were similar to the findings of other authors.19,31–34 The cleavage of partial sequences of the 16S region of the ribosomal DNA allowed for differentiation of the genus Bacillus (3918, S1, and S2) and Pseudomonas (MF2, MF4, and Ca) from other strains (CIIb, R1, and FL2). However, the observed restriction profiles did not differ among strains S1, S2, and 3918 (the species of B. subtilis) or among MF2, MF4, and Ca (the species of Pseudomonas), most likely because the 16S region of the ribosomal DNA of these species was highly conserved. Tassa and Duarte20 were also not successful in separating Pectobacterium carotovorum subsp. brasiliensis, P. carotovorum subsp. carotovorum, and the other pectobacteria available in GenBank due to the low specificity presented by the enzymatic digestion of the amplified recA gene. However, cleavage with the HhaI and TasI enzymes allowed for the separation of 13 different groups and the discrimination of P. carotovorum subsp. brasiliensis.
The same molecular method could be used to separate B. subtilis from the Pseudomonas strains, but it requires amplification of the ITS rDNA regions. Because mutations in this region are more frequent than in the ribosomal genes, which are very useful for intraspecific separation.29,35 The intergenic space (ITS) region has been used to differentiate individuals, including genetically related species.29,30,35 For example, Xanthomonas axonopodis pv. citri Type A can be differentiated from X. axonopodis pv. aurantifolii Types B and C and X. axonopodis pv. citrumela Type E by amplifying the ITS region between the 16S and 23S regions, followed by posterior cleavage with the DdeI, AluI, and Sau3AI restriction enzymes.30
ConclusionsMorphological characterization of the colonies, assessment of antibiotic sensitivity, and PCR-RFLP analysis can be used to separate the nine strains of rhizobacteria tested.
The concomitant use of the three methods presented in this work reduces the likelihood of a contaminant being multiplied with the selected rhizobacteria strain.
Conflicts of interestThe authors declare no conflicts of interest.
This research was supported by CNPq, FAPEMIG and CAPES.