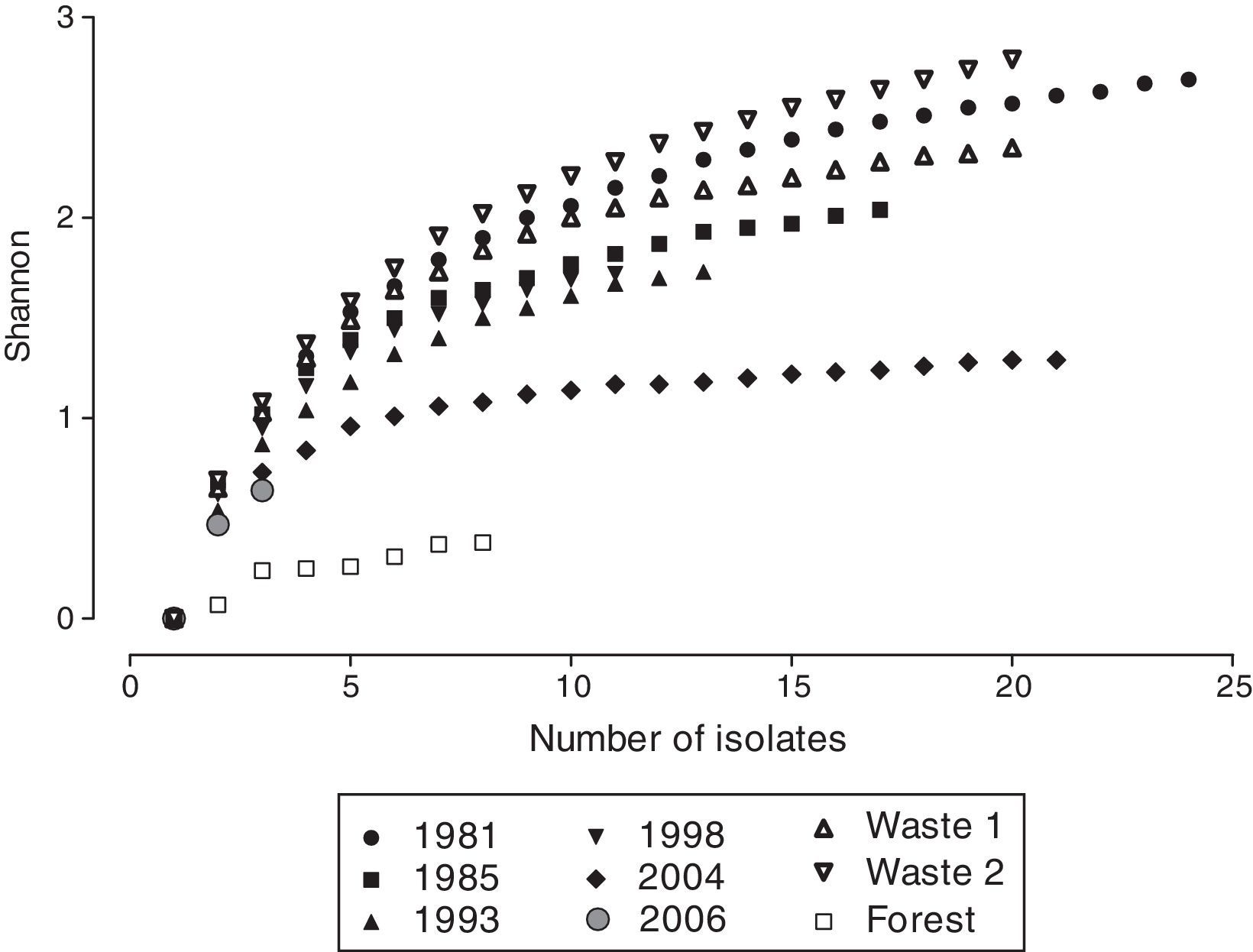
Little is known regarding how the increased diversity of nitrogen-fixing bacteria contributes to the productivity and diversity of plants in complex communities. However, some authors have shown that the presence of a diverse group of nodulating bacteria is required for different plant species to coexist. A better understanding of the plant symbiotic organism diversity role in natural ecosystems can be extremely useful to define recovery strategies of environments that were degraded by human activities. This study used ARDRA, BOX-PCR fingerprinting and sequencing of the 16S rDNA gene to assess the diversity of root nodule nitrogen-fixing bacteria in former bauxite mining areas that were replanted in 1981, 1985, 1993, 1998, 2004 and 2006 and in a native forest. Among the 12 isolates for which the 16S rDNA gene was partially sequenced, eight, three and one isolate(s) presented similarity with sequences of the genera Bradyrhizobium, Rhizobium and Mesorhizobium, respectively. The richness, Shannon and evenness indices were the highest in the area that was replanted the earliest (1981) and the lowest in the area that was replanted most recently (2006).
Nitrogen-fixing bacteria are an extremely important group of microorganisms for various ecosystems because they promote the entry of nitrogen into the soil. The capacity to fix atmospheric nitrogen is widely distributed among microorganisms with different levels of phylogenetic relationships, including representatives of Archaea and Eubacteria. However, the capacity to fix atmospheric nitrogen and induce nodule formation in leguminous plants is restricted to members of the proteobacteria phylum.1–4 Legume nodulating nitrogen-fixing bacteria, which are commonly known as rhizobia, are abundant in the soil of many ecosystems5 and have a high diversity and variability regarding symbiotic efficiency.6–8
The importance of symbiotic biological nitrogen fixation (BNF) in agricultural systems is well-documented in plant species such as soybeans, common beans and peanuts.6,9,10 However, the role of this group of microorganisms in natural ecosystems is poorly understood.11,12 Little is known about the contribution of the increased diversity of nitrogen-fixing bacteria to the productivity and diversity of plants living in natural communities. Melloni et al.13 reported that a greater diversity of bacteria in the soil results in greater resilience of the system and that a higher diversity of legume nodulating bacteria can favor symbiosis with various leguminous plant species and maximize the biological fixation of nitrogen in degraded areas. Previous research by van der Heijden et al.11 demonstrated that symbiotic nitrogen-fixing bacteria promote evenness, productivity and nitrogen capture in systems that are rich in leguminous species, which suggests that the presence of nodulating bacteria is necessary for different species of leguminous and non-leguminous plants to coexist.
Although mining activities generally alter a proportionally smaller area than other human activities, such as farming and planting pastures for livestock, the level of environmental degradation is very high because of the intense disturbance of the soil. This makes it necessary to take measures to restore these degraded areas at the end of the mining operations. In Brazil, to promote the rapid revegetation of highly degraded mined areas, the planting of leguminous species inoculated with nitrogen-fixing bacteria and arbuscular mycorrhizal fungi has been successfully employed.14 The planting of leguminous species with selected isolates of these microorganisms enables the initial colonization of substrates that have been subjected to high chemical, physical and biological degradation.15 The colonization with legumes leads to the deposition of litter and increases the concentrations of nutrients in the soil surface, enabling the replanted sites to enter the initial stages of plant succession.15
Within this context and in this study, we assessed the diversity of these bacteria in areas that were revegetated after bauxite mining to better understand their role in degraded ecosystems under the recovery process. The areas studied were revegetated between 1981 and 2006 on soil consisting of overburden or tailings and used mixes of native species and inoculated leguminous species.
Materials and methodsStudy area and collection procedureThe company Mineração Rio do Norte (MRN) operates the Saracá, Almeidas and Avisos mines (all within the Saracá-Taqüera National Forest, which is located in the municipality of Oriximiná, Pará state/Brazil, at 1°21′S – 56°22′W, 180m elevation).16 In these mines, ore is found at an average depth of 8m and is covered by dense vegetation and a layer called overburden, which is composed of organic soil, nodular bauxite and ferruginous laterite. To mine the reserves, it is necessary to remove the overburden to reveal the economically exploitable bauxite ore. This operation is conducted sequentially in which the overburden is deposited in an adjacent pit that was previously mined. In these areas, the replanting is performed on the overburden. The bauxite ore is crushed, cycloned and filtered. At the end of this process, 27% of the solid mass is tailings, which are deposited in ponds.16 These tailings pond areas are then revegetated when they become full.
The replanting of the overburden areas investigated in this study was conducted by a company using available seeds of various species (Parkia multijiga, Parkia pendula, Parkia oppositifolia, Ormosia holerythra, Ormosia excelsa, Sclerolobium paniculatum and Acosmium nitens) in 1981, 1985, 1993, 1998, 2004 and 2006. The revegetation of the two tailings ponds was conducted using the species Acacia mangium in 1993. In part of this area, the planting was conducted without rhizobia inoculation (Tailings Waste 1), and the other part with the rhizobia inoculation consisted of a mixture of all of the recommended rhizobial strains from Embrapa Agrobiologia (Tailings Waste 2).
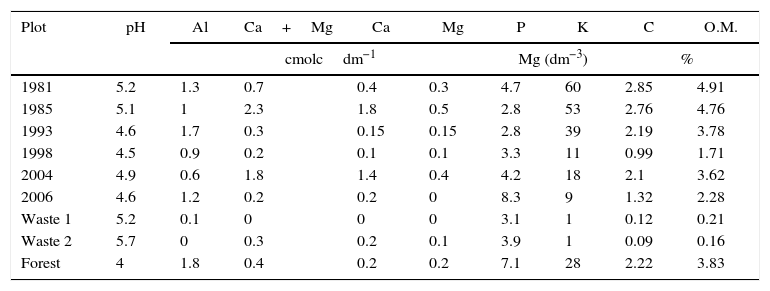
In each area, twenty simple samples (0–0.20m) were harvested to compose a compound sample in plots measuring 250m2. Nine compound samples were collected in 2007: six in plots that had been revegetated on overburden, two in tailings plots and one in a native forest plot (Table 1). The chemical analysis of the soil samples was conducted according to the Manual on Soil Analysis Methods.17
Chemical characteristics of the soil samples.
| Plot | pH | Al | Ca+Mg | Ca | Mg | P | K | C | O.M. |
|---|---|---|---|---|---|---|---|---|---|
| cmolcdm−1 | Mg (dm−3) | % | |||||||
| 1981 | 5.2 | 1.3 | 0.7 | 0.4 | 0.3 | 4.7 | 60 | 2.85 | 4.91 |
| 1985 | 5.1 | 1 | 2.3 | 1.8 | 0.5 | 2.8 | 53 | 2.76 | 4.76 |
| 1993 | 4.6 | 1.7 | 0.3 | 0.15 | 0.15 | 2.8 | 39 | 2.19 | 3.78 |
| 1998 | 4.5 | 0.9 | 0.2 | 0.1 | 0.1 | 3.3 | 11 | 0.99 | 1.71 |
| 2004 | 4.9 | 0.6 | 1.8 | 1.4 | 0.4 | 4.2 | 18 | 2.1 | 3.62 |
| 2006 | 4.6 | 1.2 | 0.2 | 0.2 | 0 | 8.3 | 9 | 1.32 | 2.28 |
| Waste 1 | 5.2 | 0.1 | 0 | 0 | 0 | 3.1 | 1 | 0.12 | 0.21 |
| Waste 2 | 5.7 | 0 | 0.3 | 0.2 | 0.1 | 3.9 | 1 | 0.09 | 0.16 |
| Forest | 4 | 1.8 | 0.4 | 0.2 | 0.2 | 7.1 | 28 | 2.22 | 3.83 |
The experiment was conducted using sterilized Leonard jars containing sand and vermiculite at a proportion of 1:1 (v/v) and in a randomized block design with three repetitions. The treatments consisted of the inoculation of a suspension of soil from each plot on the host trap plants Macroptilium atropurpureum (siratro) and Mimosa acutistipula.
The seeds were treated with concentrated sulfuric acid (H2SO4) for 10min to break the dormancy and with 30% hydrogen peroxide (H2O2) for 3min for surface disinfection. They were then germinated in Petri dishes containing moistened filter paper and cotton. Three seedlings of each species were transplanted to each jar, and each seedling was inoculated with 1mL of the soil suspension in saline solution. The suspension was prepared using 10g of soil in 90mL of NaCl solution (0.145M) and kept under orbital agitation for 30min.
The experiment lasted 90 days, during which the plants received water and a nutrient solution18 intercalated every 15 days. To isolate the bacteria present in the nodules, they were washed in ethanol (70%, v/v – 1min), externally disinfected with 30% hydrogen peroxide for 3min, washed five times in sterile distilled water and crushed in Petri dishes containing YMA medium.19
Molecular characterizationThe restriction analysis of the 16S rDNA gene was conducted according to Laguerre et al.20 and Teixeira et al.8 using HinfI, MspI and DdeI endonucleases. The DNA was extracted according to Doyle and Doyle21 using the detergent CTAB. The 16S rDNA gene was amplified using the universal primers Y1 (5′-TGGCTCAGAACGAACGCTGGCGGC-3′) and Y3 (5′-CTGACCCCACTTCAGCATTGTTCCAT-3′).22
We selected isolates representing 12 distinct clusters in the ARDRA dendrogram for partial sequencing of the 16S rDNA gene. The PCR product was purified using the Wizard® SV Gel and PCR Clean-Up System kit (Promega Corporation, Madison, WI, USA) following the manufacturer's recommendations. The reactions were conducted with the DYEnamic™ ET Dye Terminator kit (MegaBACE™) and a MegaBACE 1000 automatic sequencer (GE Healthcare Life Sciences). The identities of the sequences (access numbers KT29901–KT429912) were estimated with the database of the National Center for Biotechnology Information through its Basic Local Alignment Search Tool.23
The BOX-PCR reactions were conducted according to Kaschuk et al.24 by employing 50ng of template DNA and the BOX-A1R primer (5′-CTACGGCAAGGCGACGCTGACG-3′).25 The similarity dendrogram was constructed using the Jaccard similarity index and the UPGMA. We assigned operational taxonomic units (OTUs) based on distinct clusters in the tree, which we then used to calculate the richness, Shannon and evenness index values by employing the PAST program and the rarefaction analysis using the Estimates version 8.0 program (Colwell, 2010)26 according to Magurran.27
ResultsIsolation of the bacteriaA total of 139 isolates were obtained and separated by origin into 24, 17, 13, 11, 22, 3, 8, 21 and 20 isolates obtained from the areas replanted in 1981, 1985, 1993, 1998, 2004, and 2006, and the Forest Area, Tailings Waste 1 and Tailings Waste 2, respectively. All of these isolates were obtained from siratro nodules. No nodulation was observed in M. acutistipula.
ARDRA and sequencingOf the 139 isolates obtained, two did not grow in the culture medium after the storage period and were excluded from the molecular analysis (one from the area replanted in 2004 and one from Tailings Waste 2), and for two other isolates (from Tailings Waste 1), no PCR product was obtained from the 16S rDNA gene to conduct the restriction. Therefore, the ARDRA was conducted with 135 isolates.
The isolates were distributed in 25 different clusters (thickest lines in Fig. 1), and the number of isolates per cluster ranged from 1 to 17. Fifty-six isolates positioned in different genetic clusters were not discriminated because they had 100% similarity. Among the 12 isolates from which the 16S gene was partially sequenced, similarity could be detected in the sequences of three genera. Of these, eight belonged to the Bradyrhizobium genus, three to Rhizobium and one to Mesorhizobium. Of all of the 135 isolates analyzed from the 16S gene restriction, 96 isolates were clustered with the Bradyrhizobium genus, 25 with Rhizobium and 8 with Mesorhizobium (Fig. 1).
BOX-PCR and diversity analysisThe BOX analysis was conducted with 137 isolates. These isolates were distributed in 40 clusters (Fig. 2), and the number of isolates per cluster varied between 1 and 12. Twelve isolates positioned in different genetic clusters were not discriminated because they had 100% similarity. Based on the generated dendrogram, we calculated the diversity of the isolates for each plot. The richness of the OTUs varied between 2 for the forest area and 17 for the area that was revegetated in 1981 and Tailings Waste 2; the Shannon index ranged from 0.37 for the forest area to 2.78 for Tailings Waste 2; and the evenness index varied between 0.54 for the forest area to 0.98 for Tailings Waste 2 (Figs. 3 and 4).
Several studies have demonstrated that species belonging to the Mimosa genus have a high specificity for rhizobia isolates belonging to the beta proteobacteria subdivision, notably, isolates of the Burkholderia genus.28,29 In addition, some species of Mimosoideae must associate with arbuscular mycorrhizal fungi for nodulation to occur.30 The absence or low density of beta-rhizobia in the sampled areas and the absence of arbuscular mycorrhizal fungi in the Leonard jars used in this study can be possible explanations for the lack of nodulation in M. acutistipula. However, the majority of siratro plants showed nodulation, which demonstrates that the density of rhizobia present in the inoculum was sufficient to induce nodulation. This species, because of its wide host range, has been previously used to assess the diversity of isolates from nodules.31,32
ARDRA was initially proposed as a useful tool for the rapid identification of the taxonomic position of rhizobia isolates because it is based on the restriction of a gene that codes for ribosomal RNA, which permits separation at the genus level and at the species level in some cases. It is mainly employed for this purpose in situations where the 16S gene cannot be sequenced.8,20,33,34 In the present study, the majority of the isolates presented sequences close to those of the Bradyrhizobium genus (groups 10–22, Fig. 1). A greater abundance of Bradyrhizobium isolates have been observed by other authors under a variety of conditions,5,32,35 which suggests that there is high variability within this genus in the areas studied. In addition to the isolates proximately related to the Bradyrhizobium genus, we also observed isolates proximately related to the Rhizobium and Mesorhizobium genera. Previous results showed the capacity of isolates belonging to these genera to withstand adverse conditions of pH, temperature and the presence of toxic elements.36–38 The presence of these genera in the areas studied confirms their capacity to survive and establish symbiosis fixing-nitrogen under adverse environmental conditions.
The BOX-PCR analysis allows the simultaneous evaluation of distinct genomic regions to identify intraspecific variability. This feature was demonstrated in this study because the isolates were distributed into 25 clusters by the ARDRA and 40 clusters by the BOX-PCR analysis. Using ARDRA, even based on data from three restriction enzymes, 56 isolates positioned in different genetic clusters showed 100% similarity, whereas this number was only 12 for the BOX-PCR technique. This is because the 16S rDNA gene is relatively small and presents highly conserved regions. Additionally, this gene is poorly discriminated within the genus Bradyrhizobium.39 These findings show that the BOX-PCR technique was more discriminating between isolates, demonstrating the applicability of this tool to separate isolates that are taxonomically proximate 40–43 and its utility in studies aiming to compare the level of diversity between different sites.
Among the areas that were revegetated on overburden, the OTU richness, Shannon and evenness indices were higher in the area that was revegetated the earliest (1981). The area revegetated more recently (2006) showed a low number of isolates, which impaired the diversity estimate because it was not possible to perceive stabilization of the Shannon index (Fig. 4) when presented with the lowest OTU richness, Shannon and evenness indices value. The other areas presented intermediate values (Fig. 3). The forest area showed OTU richness, Shannon and evenness indices lower than all of the overburden areas, and the two tailings waste areas that were revegetated using Acacia species presented similar indices to the overburden area revegetated in 1981 and higher indices than in the overburden area that was revegetated in the same year (1993) using a mixture of plant species.
A lower diversity of nodule isolates in forest areas compared cultivated areas or fallows has been previously reported in several studies.13,30,32 Jesus et al.31 evaluated the effect of the type of land use on the diversity of nodule isolates captured by siratro and observed that the area cultivated with cassava presented a higher diversity than the forest areas and those cultivated with peach palm, which did not differ between one another. Lima et al.32 also investigated the community of nodule isolates from siratro in areas with different uses in the Amazon region and observed that the richness index of the nodule isolates in the primary forest area was 12, which was the lowest value among the areas analyzed. The values in the other areas were 46 for the cultivated area, 48 in the agroforestry area, 24 for the area identified as old secondary forest, 28 in new secondary forest and 29 in the pasture.
These results show that the revegetation strategies used in these areas enabled the establishment of a plant community that was able to sustain an increasing diversity of root nodule isolates. According to Melloni et al., 13 studies of the diversity of key groups of microorganisms in reclaimed areas are important because they supply an indication of the effects of different rehabilitation methods on the diversity of these microorganisms.
The tailings pond areas were replanted with Acacia mangium in 1993. Since then, the changes caused by the coverages with leguminous species have led to the deposition of plant material on the soil, which provided conditions for the establishment of a diverse community of root nodule isolates. A higher diversity of legume-nodulating and nitrogen-fixing bacteria was also found in areas that were replanted with the leguminous species Mimosa scabrella and Cajanus cajan after mining activities.13 The capacity of leguminous species to colonize degraded environments has been reported by other authors 14,15 and has been linked to the ability of these species form symbiotic associations with nitrogen-fixing bacteria in the soil, as well as with arbuscular mycorrhizal fungi. This three-way interaction favors the nutrition of plants by enhancing their uptake of nutrients. As a result of the increase of nitrogen in the biomass and the absorption of important nutrients, such as phosphorous, leguminous plants favor plant succession and the diversity of microorganisms.
Conflicts of interestThe authors declare no conflicts of interest.
We would like to thank Mineração Rio do Norte for allowing access to the study area; the National Council for Scientific and Technological Development (CNPq) for individual support (Borges, W.L. received a doctoral scholarship, process no. 142315/2007–9; de Faria, S.M. received a research grant) and project funding (no. 492683/2004-2), and IBAMA (029/2007 02001.006557/2005) and the Graduate Program in Agronomy – Soil Science of UFRRJ and Embrapa for the infrastructure that was made available. We would also like to thank Telmo Felix da Silva, Carlos Fernando da Cunha, Cândido Barreto de Novais and Adriana Santos do Nascimento for help in the greenhouse experiment and with the bacterial isolation.