The present work discusses the technological and new selection criteria that should be included for selecting lactic acid bacteria for production of fermented meat. Lactic acid bacteria isolated from Bulgarian traditional fermented “lulanka” salami was studied regarding some positive technological parameters (growth at different temperature, pH, and proteolytic activity). The presence of genes related to the virulence factors, production of biogenic amines, and vancomycin resistance were presented in low frequency in the studied lactic acid bacteria. On the other hand, production of antimicrobial peptides and high spread of bacteriocin genes were broadly presented. Very strong activity against L. monocytogenes was detected in some of the studied lactic acid bacteria. In addition, the studied strains did not present any antimicrobial activity against tested closely related bacteria such as Lactobacillus spp., Lactococcus spp., Enterococcus spp. or Pediococcus spp. To our knowledge this is the first study on the safety and antimicrobial properties of lactic acid bacteria isolated from Bulgarian lukanka obtained by spontaneous fermentation.
The production of fermented meat products is part of the gastronomic habits of human culture from all continents and can be traced back to earliest records dating from 1500 BC. Besides the improved preservation characteristics and safety of the final product, fermentation processes play an essential role in the organoleptic properties of meat products. This role is related to the presence and interactions between different starter cultures and autochthonous microbiota found in products of animal origin, different additives of both an organic and inorganic nature, essential oils, non digestive supplements, fibres, and specific metabolitic products. Consumer demands for high-quality, safe, nutritious, and convenient meat products have provided important insights into the development of a host of processed meat products, including fermented meats. This offers unique perspectives for the application of novel functional microbial cultures and promising emerging technologies in the manufacture of fermented meats. Fermented meat products are unique and often represented as an element of culinary heritage and identity.
Different geographical areas developed unique varieties of preserved meat products that varied in size, shape, texture, appearance, and flavour.1 There has been a renewed interest in traditional fermented meat products, mainly in Europe, where they have a great significance and economic impact.2 The fermentation of meat products is related to a number of physical, biochemical and microbial changes in the muscle-based product, caused by endogenous and microbial enzymatic activities. The taste of fermented meat products is primarily due to the presence of lactic acid and the production of low molecular weight flavour compounds, such as peptides, free amino acids, aldehydes, organic acids, and amines, arising from the proteolysis of meat.3 Natural fermentation of meat can also affect the uniformity of products, which depend on the specific composition of the so-called ‘houseflora’. For all of these reasons, the addition of starter cultures has been recommended for the manufacture of fermented meat. The use of starter cultures has become increasingly necessary in order to produce a more consistent and stable product by improving quality, reducing variability and enhancing the organoleptic characteristics of fermented meat products.4,5 In addition novel starter cultures may include microorganisms that generate health-promoting molecules, possess probiotic attributes or reduce the level of biogenic amine and cholesterol content in fermented meat products.4 In addition to all these, the safety of the starter cultures used is an essential issue. Probiotics are live microorganisms which, when ingested in sufficient quantities, can exert some beneficial health effects in the host, and are primarily derived from Bifidobacterium and Lactobacillus species. Additionally, some species of Lactococcus, Enterococcus, Saccharomyces, and Propionibacterium were also reported to present probiotic characteristics.6 Several studies have demonstrated the feasibility of adding probiotic cultures to fermented meat product formulations. Erkkila et al.7 examined the ability of three probiotic Lactobacillus rhamnosus strains to be involved in the production of fermented dry sausage; Pennacchia et al.8 reported the use of Lactobacillus plantarum and Lactobacillus paracasei as probiotics in meat products; Pediococcus acidilactici and Lactobacillus sakei also showed good survival characteristics in fermented sausages; both are considered as being good probiotic candidates for use in meat products9; Todorov et al.10 investigated the safety aspects of Lb. sakei, Lb. plantarum, and Enterococcus faecium strains isolated from Portuguese fermented meat products in order to apply them as a beneficial (starter, bacteriocin producers or probiotics) culture. The bacteriocins produced by starter cultures may have an advantage for these strains in competitive interactions with the pathogenic bacteria from the food matrix and can be considered as being advantageous for the probiotic cultures as well.
Lukanka is a Bulgarian (sometimes spicy) salami which is unique to Bulgarian cuisine. Lukanka is semi-dried, has a flattened cylindrical shape, and brownish-red interior in a skin that is normally covered with a white fungus. The final product is characterized as having low water activity, slightly acid taste, and can be stored under refrigeration (during summer) or at the environmental temperature in the winter period. The mix of small pieces of meat and fat give the interior a grainy structure. Traditionally, lukanka salami is made of pork, veal, and spices (black pepper, cumin, salt), minced together and stuffed into a length of dried cow's intestine as casing. After the stuffing process, the cylindrical salami is hung to dry for about 40–50 days in a well-ventilated location. In the process of drying, the salami is pressed to acquire its typical flat form.
In the preliminary works, more than 200 isolates of the genera Lactobacillus were isolated during the different stages of the fermentation process of the naturally fermented lukanka.12 The isolates were taxonomic identified by 16S rRNA and the majority was classified as being related to Lb. plantarum, Lb. brevis and Lb. sakei. Research into the production of lactic acid, determination of pH and screening for producers of antimicrobials was performed. One of the most important roles of Lactobacillus strains in this process is their acidifying properties for the preservation of the final product, depending as well on the total cell number, and of the acidifying properties of the particular strains.12
The aim of this work was to investigate beneficial, technological properties, and safety aspects of lactic acid bacteria (LAB) isolated from “lukanka”, including growth at different temperature, pH, and proteolytic activity; presence of genes related to virulence factors, production of biogenic amines, and vancomycin resistance; production of bacteriocins and presence of related genes.
Materials and methodsStrains and mediaLb. plantarum (13 strains: S2, S3, S5, S7, S9, S18, S20, S23, S29, S30, S35, S37, and S39), Lb. brevis (4 strains: S8, S27, S36 and S38) and Lb. sakei (4 strains: S11, S12, S13 and S15) isolated from the Bulgarian dry sausage lukanka, produced by spontaneous fermentation (without adding starter cultures) were previously isolated, identified and some of them characterized as bacteriocin producers and cultured at 30°C in MRS broth (Difco, Le Pont de Claix, France).12Lactobacillus spp., Lactococcus spp., Enterococcus spp., Pediococcus spp. and L. monocytogenes strains used in the bacteriocin production test as test microorganisms (Table 1) were grown on MRS (Difco) at 30°C and BHI (Difco) at 37°C, respectively, and stored in the presence of 20% glycerol at −80°C. Before use, the strains were cultivated at least twice in MRS or BHI media.
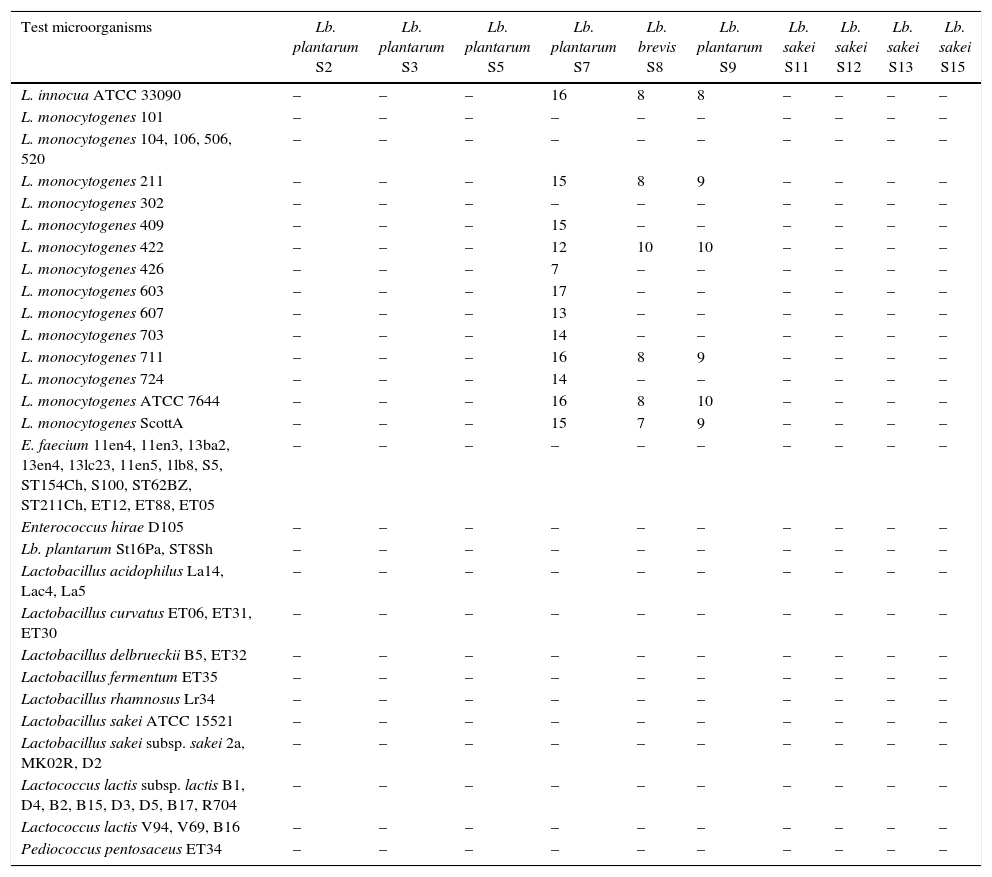
Antimicrobial spectrum of activity of selected LAB isolated from “Lukanka”.
| Test microorganisms | Lb. plantarum S2 | Lb. plantarum S3 | Lb. plantarum S5 | Lb. plantarum S7 | Lb. brevis S8 | Lb. plantarum S9 | Lb. sakei S11 | Lb. sakei S12 | Lb. sakei S13 | Lb. sakei S15 |
|---|---|---|---|---|---|---|---|---|---|---|
| L. innocua ATCC 33090 | – | – | – | 16 | 8 | 8 | – | – | – | – |
| L. monocytogenes 101 | – | – | – | – | – | – | – | – | – | – |
| L. monocytogenes 104, 106, 506, 520 | – | – | – | – | – | – | – | – | – | – |
| L. monocytogenes 211 | – | – | – | 15 | 8 | 9 | – | – | – | – |
| L. monocytogenes 302 | – | – | – | – | – | – | – | – | – | – |
| L. monocytogenes 409 | – | – | – | 15 | – | – | – | – | – | – |
| L. monocytogenes 422 | – | – | – | 12 | 10 | 10 | – | – | – | – |
| L. monocytogenes 426 | – | – | – | 7 | – | – | – | – | – | – |
| L. monocytogenes 603 | – | – | – | 17 | – | – | – | – | – | – |
| L. monocytogenes 607 | – | – | – | 13 | – | – | – | – | – | – |
| L. monocytogenes 703 | – | – | – | 14 | – | – | – | – | – | – |
| L. monocytogenes 711 | – | – | – | 16 | 8 | 9 | – | – | – | – |
| L. monocytogenes 724 | – | – | – | 14 | – | – | – | – | – | – |
| L. monocytogenes ATCC 7644 | – | – | – | 16 | 8 | 10 | – | – | – | – |
| L. monocytogenes ScottA | – | – | – | 15 | 7 | 9 | – | – | – | – |
| E. faecium 11en4, 11en3, 13ba2, 13en4, 13lc23, 11en5, 1lb8, S5, ST154Ch, S100, ST62BZ, ST211Ch, ET12, ET88, ET05 | – | – | – | – | – | – | – | – | – | – |
| Enterococcus hirae D105 | – | – | – | – | – | – | – | – | – | – |
| Lb. plantarum St16Pa, ST8Sh | – | – | – | – | – | – | – | – | – | – |
| Lactobacillus acidophilus La14, Lac4, La5 | – | – | – | – | – | – | – | – | – | – |
| Lactobacillus curvatus ET06, ET31, ET30 | – | – | – | – | – | – | – | – | – | – |
| Lactobacillus delbrueckii B5, ET32 | – | – | – | – | – | – | – | – | – | – |
| Lactobacillus fermentum ET35 | – | – | – | – | – | – | – | – | – | – |
| Lactobacillus rhamnosus Lr34 | – | – | – | – | – | – | – | – | – | – |
| Lactobacillus sakei ATCC 15521 | – | – | – | – | – | – | – | – | – | – |
| Lactobacillus sakei subsp. sakei 2a, MK02R, D2 | – | – | – | – | – | – | – | – | – | – |
| Lactococcus lactis subsp. lactis B1, D4, B2, B15, D3, D5, B17, R704 | – | – | – | – | – | – | – | – | – | – |
| Lactococcus lactis V94, V69, B16 | – | – | – | – | – | – | – | – | – | – |
| Pediococcus pentosaceus ET34 | – | – | – | – | – | – | – | – | – | – |
| Test microorganisms | Lb. plantarum S18 | Lb. plantarum S20 | Lb. plantarum S23 | Lb. brevis S27 | Lb. plantarum S29 | Lb. plantarum S30 | Lb. plantarum S35 | Lb. brevis S36 | Lb. plantarum S37 | Lb. brevis S38 | Lb. plantarum S39 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| L. innocua ATCC 33090 | – | 10 | – | – | – | 16 | – | 17 | 14 | – | 17 |
| L. monocytogenes 101 | – | – | – | – | – | – | – | – | 17 | – | 18 |
| L. monocytogenes 104, 106, 506, 520 | – | – | – | – | – | – | – | – | – | – | – |
| L. monocytogenes 211 | – | – | – | – | – | 10 | – | 11 | 15 | – | 17 |
| L. monocytogenes 302 | – | 8 | – | – | – | 9 | – | 7 | – | – | – |
| L. monocytogenes 409 | – | – | – | – | – | – | – | – | 12 | – | 13 |
| L. monocytogenes 422 | – | – | – | – | – | 8 | – | 7 | 10 | – | 12 |
| L. monocytogenes 426 | – | – | – | – | – | – | – | – | – | – | – |
| L. monocytogenes 603 | – | – | – | – | – | 8 | – | 9 | 16 | – | 17 |
| L. monocytogenes 607 | – | – | – | – | – | 11 | – | 12 | 10 | – | 8 |
| L. monocytogenes 703 | – | – | – | – | – | 7 | – | 7 | – | – | – |
| L. monocytogenes 711 | – | – | – | – | – | 9 | – | – | 11 | – | 14 |
| L. monocytogenes 724 | – | – | – | – | – | 13 | – | 12 | 10 | – | 11 |
| L. monocytogenes ATCC 7644 | – | 9 | – | – | – | 14 | – | 15 | 17 | – | 18 |
| L. monocytogenes ScottA | – | – | – | – | 17 | – | 16 | 16 | – | 17 | |
| E. faecium 11en4, 11en3, 13ba2, 13en4, 13lc23, 11en5, 1lb8, S5, ST154Ch, S100, ST62BZ, ST211Ch, ET12, ET88, ET05 | – | – | – | – | – | – | – | – | – | – | – |
| Enterococcus hirae D105 | – | – | – | – | – | – | – | – | – | – | – |
| Lb. plantarum St16Pa, ST8Sh | – | – | – | – | – | – | – | – | – | – | – |
| Lactobacillus acidophilus La14, Lac4, La5 | – | – | – | – | – | – | – | – | – | – | – |
| Lactobacillus curvatus ET06, ET31, ET30 | – | – | – | – | – | – | – | – | – | – | – |
| Lactobacillus delbrueckii B5, ET32 | – | – | – | – | – | – | – | – | – | – | – |
| Lactobacillus fermentum ET35 | – | – | – | – | – | – | – | – | – | – | – |
| Lactobacillus rhamnosus Lr34 | – | – | – | – | – | – | – | – | – | – | – |
| Lactobacillus sakei ATCC 15521 | – | – | – | – | – | – | – | – | – | – | – |
| Lactobacillus sakei subsp. sakei 2a, MK02R, D2 | – | – | – | – | – | – | – | – | – | – | – |
| Lactococcus lactis subsp. lactis B1, D4, B2, B15, D3, D5, B17, R704 | – | – | – | – | – | – | – | – | – | – | – |
| Lactococcus lactis V94, V69, B16 | – | – | – | – | – | – | – | – | – | – | – |
| Pediococcus pentosaceus ET34 | – | – | – | – | – | – | – | – | – | – | – |
Antimicrobial activity was presented as diameter of the inhibition zones in millimetres; –, no inhibition zone.
Growth of Lb. plantarum, Lb. brevis and Lb. sakei strains was determined in MRS broth in the presence of NaCl (2%, 4%, 6.5%, 10% and 18% (m/v)). In addition, strains were cultured at 15°C, 25°C, 37°C, 40°C and 50°C in MRS broth. The ability of the studied strains to develop in the medium at different initial pH (pH 3.0–pH 5.0) was monitored during their cultivation in MRS, as well as their growth capability on acetate agar with pH 5.4 (Sigma–Aldrich, Co, St. Luis, MO, USA).
Growth at 37°C in MRS broth (recommended optimal conditions for LAB) was accepted as baseline criteria and referred as “copious”. When growth parameters of investigated LAB were lower than 30% compared to optimal (copious), a bacterial growth was defined as “moderate”. Growth parameters lower than 70% compared to baseline were referred as “abundant”.
Detection of proteolitic activitySDS-PAGE was used for determination of the proteolitic activity of the investigated strains.
Production of enzymesThe enzymatic activity of the studied was determined using APIZYM strips (bioMérieux, Marcy-l’Etoile, France) according to the manufacturer's instructions. The 20 (1 control and 19 different substrates, Table 2) variants from the APIZYM strips were inoculated with 24-h-old cultures grown on MRS agar (Difco), and then incubated at 30°C for 4h. The evaluation of the activity was graded from 0 to 5 using the APIZYM colour reaction chart according to the intensity of colouration and manufacturer's instructions.
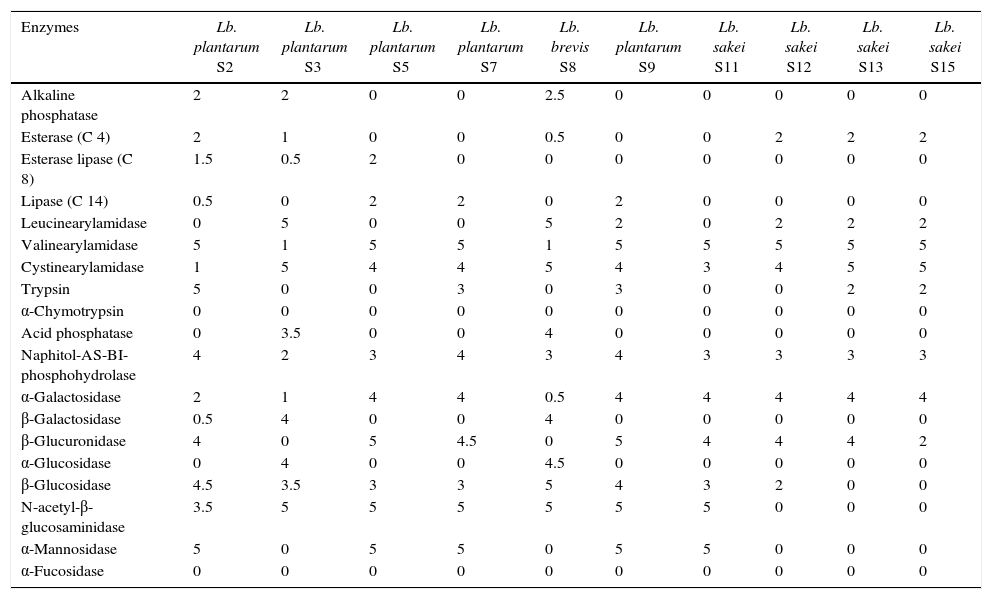
API ZYM enzymatic profile of LAB isolated from “Lukanka”.
| Enzymes | Lb. plantarum S2 | Lb. plantarum S3 | Lb. plantarum S5 | Lb. plantarum S7 | Lb. brevis S8 | Lb. plantarum S9 | Lb. sakei S11 | Lb. sakei S12 | Lb. sakei S13 | Lb. sakei S15 |
|---|---|---|---|---|---|---|---|---|---|---|
| Alkaline phosphatase | 2 | 2 | 0 | 0 | 2.5 | 0 | 0 | 0 | 0 | 0 |
| Esterase (C 4) | 2 | 1 | 0 | 0 | 0.5 | 0 | 0 | 2 | 2 | 2 |
| Esterase lipase (C 8) | 1.5 | 0.5 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Lipase (C 14) | 0.5 | 0 | 2 | 2 | 0 | 2 | 0 | 0 | 0 | 0 |
| Leucinearylamidase | 0 | 5 | 0 | 0 | 5 | 2 | 0 | 2 | 2 | 2 |
| Valinearylamidase | 5 | 1 | 5 | 5 | 1 | 5 | 5 | 5 | 5 | 5 |
| Cystinearylamidase | 1 | 5 | 4 | 4 | 5 | 4 | 3 | 4 | 5 | 5 |
| Trypsin | 5 | 0 | 0 | 3 | 0 | 3 | 0 | 0 | 2 | 2 |
| α-Chymotrypsin | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Acid phosphatase | 0 | 3.5 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 |
| Naphitol-AS-BI-phosphohydrolase | 4 | 2 | 3 | 4 | 3 | 4 | 3 | 3 | 3 | 3 |
| α-Galactosidase | 2 | 1 | 4 | 4 | 0.5 | 4 | 4 | 4 | 4 | 4 |
| β-Galactosidase | 0.5 | 4 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 |
| β-Glucuronidase | 4 | 0 | 5 | 4.5 | 0 | 5 | 4 | 4 | 4 | 2 |
| α-Glucosidase | 0 | 4 | 0 | 0 | 4.5 | 0 | 0 | 0 | 0 | 0 |
| β-Glucosidase | 4.5 | 3.5 | 3 | 3 | 5 | 4 | 3 | 2 | 0 | 0 |
| N-acetyl-β-glucosaminidase | 3.5 | 5 | 5 | 5 | 5 | 5 | 5 | 0 | 0 | 0 |
| α-Mannosidase | 5 | 0 | 5 | 5 | 0 | 5 | 5 | 0 | 0 | 0 |
| α-Fucosidase | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Enzymes | Lb. plantarum S18 | Lb. plantarum S20 | Lb. plantarum S23 | Lb. brevis S27 | Lb. plantarum S29 | Lb. plantarum S30 | Lb. plantarum S35 | Lb. brevis S36 | Lb. plantarum S37 | Lb. brevis S38 | Lb. plantarum S39 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Alkaline phosphatase | 1 | 0 | 0 | 3 | 0 | 0 | 0 | 1 | 2 | 3.5 | 0 |
| Esterase (C 4) | 1 | 2 | 2 | 2 | 1 | 2 | 2 | 2 | 1 | 1 | 1 |
| Esterase lipase (C 8) | 1 | 2 | 2 | 1.5 | 2 | 2 | 2 | 2 | 2 | 0.5 | 0.5 |
| Lipase (C 14) | 1 | 2 | 2 | 0 | 2 | 2 | 2 | 1 | 0 | 0 | 0 |
| Leucinearylamidase | 1 | 2 | 2 | 5 | 2 | 2.5 | 2 | 2 | 1 | 5 | 1 |
| Valinearylamidase | 5 | 5 | 5 | 2.5 | 5 | 5 | 5 | 5 | 5 | 1 | 5 |
| Cystinearylamidase | 4 | 4 | 4 | 5 | 4 | 4 | 4 | 4 | 4 | 5 | 4 |
| Trypsin | 3 | 3 | 3 | 3 | 0 | 3 | 3 | 3 | 3 | 0 | 3 |
| α-Chymotrypsin | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Acid phosphatase | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 3.5 | 0 |
| Naphitol-AS-BI-phosphohydrolase | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 2 | 3 |
| α-Galactosidase | 4 | 5 | 5 | 3 | 4 | 5 | 4 | 4 | 4 | 2 | 3 |
| β-Galactosidase | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 4 | 0 |
| β-Glucuronidase | 4 | 5 | 5 | 4.5 | 4 | 5 | 5 | 4 | 4 | 0 | 2 |
| α-Glucosidase | 0 | 0 | 0 | 3.5 | 0 | 0 | 0 | 0 | 0 | 4 | 0 |
| β-Glucosidase | 4 | 4 | 4 | 3 | 5 | 4 | 3 | 3 | 3 | 5 | 3 |
| N-acetyl-β-glucosaminidase | 5 | 5 | 5 | 4 | 5 | 5 | 5 | 5 | 5 | 5 | 5 |
| α-Mannosidase | 5 | 5 | 5 | 0 | 5 | 5 | 5 | 5 | 5 | 0 | 0 |
| α-Fucosidase | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
Selected microorganisms were screened for bacteriocin production using the agar-spot-test method against a panel of test microorganisms, including various strains of Listeria spp., Enterococcus spp., and Lactobacillus spp. (Table 2).13 Strains were grown in MRS broth for 24h at 30°C, cell-free supernatant was obtained after centrifugation (6000g, 10min, 4°C), followed by correction of pH to 6.0–6.5 with 1M NaOH and thermal treatment for 10min at 80°C. Ten microliters of previously prepared material were spotted on the surface of MRS or BHI containing 1% agar, inseminated with one of the previously mentioned test microorganisms (Table 1) at an approximate concentration of 106CFU/ml, and incubated for 24h at an optimal temperature for the test microorganism (30°C or 37°C).
Screening for presence of bacteriocin genesTotal DNA was isolated from the selected microorganisms using ZR Fungal/Bacterial DNA Kit (Zymo Research, Irvine, CA, USA) according to the manufacturer's instructions. DNA was submitted to PCR reactions to detect genes responsible for codification of the following bacteriocins: nisin, pediocin PA-1, curvacin A, plantaricin S, plantaricin NC8, plantaricin W, sakacin G, sakacin Tα, sakacin Tβ and sakacin X (Table 3).14–19 The PCR reaction was prepared using primers at 10pM/μL and conditions as described previously, adjusting the annealing temperature according to the specification of the primers used.14–19 The amplified products were separated by electrophoresis on agarose gels in 0.5× TAE buffer. Agarose gels were stained in 0.5× TAE buffer containing 0.5μg/ml ethidium bromide (Sigma).
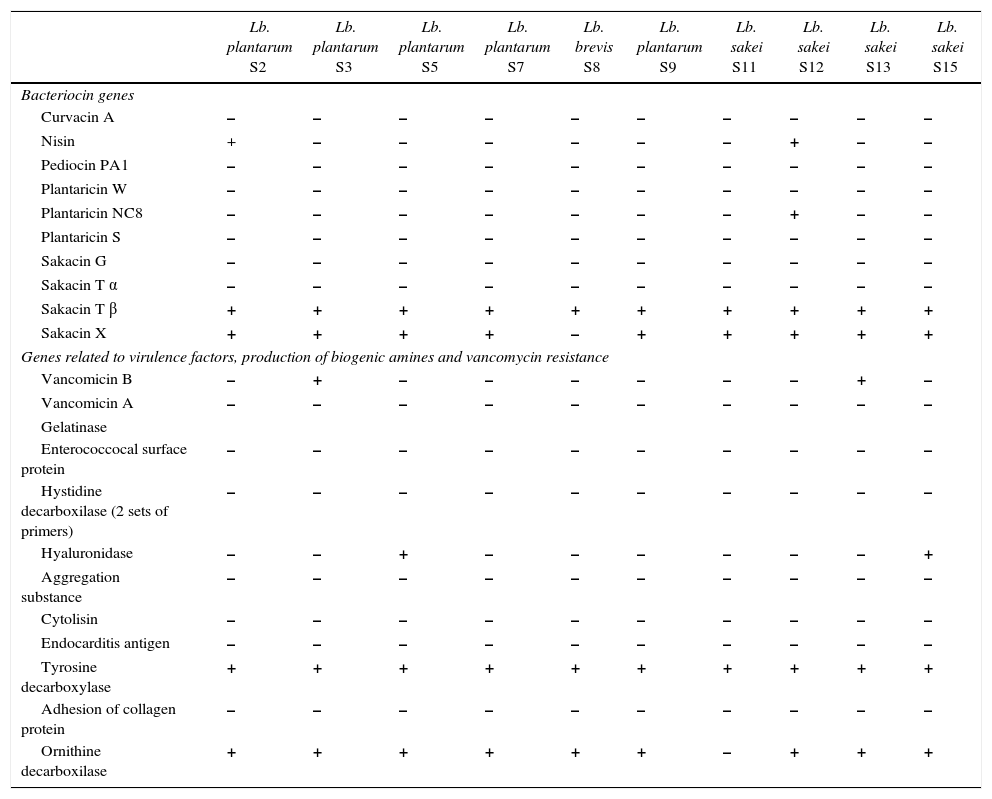
Presence of bacteriocin genes and genes related to virulence factors, production of biogenic amines and vancomycin resistance in LAB isolated from “Lukanka”.
| Lb. plantarum S2 | Lb. plantarum S3 | Lb. plantarum S5 | Lb. plantarum S7 | Lb. brevis S8 | Lb. plantarum S9 | Lb. sakei S11 | Lb. sakei S12 | Lb. sakei S13 | Lb. sakei S15 | |
|---|---|---|---|---|---|---|---|---|---|---|
| Bacteriocin genes | ||||||||||
| Curvacin A | – | – | – | – | – | – | – | – | – | – |
| Nisin | + | – | – | – | – | – | – | + | – | – |
| Pediocin PA1 | – | – | – | – | – | – | – | – | – | – |
| Plantaricin W | – | – | – | – | – | – | – | – | – | – |
| Plantaricin NC8 | – | – | – | – | – | – | – | + | – | – |
| Plantaricin S | – | – | – | – | – | – | – | – | – | – |
| Sakacin G | – | – | – | – | – | – | – | – | – | – |
| Sakacin T α | – | – | – | – | – | – | – | – | – | – |
| Sakacin T β | + | + | + | + | + | + | + | + | + | + |
| Sakacin X | + | + | + | + | – | + | + | + | + | + |
| Genes related to virulence factors, production of biogenic amines and vancomycin resistance | ||||||||||
| Vancomicin B | – | + | – | – | – | – | – | – | + | – |
| Vancomicin A | – | – | – | – | – | – | – | – | – | – |
| Gelatinase | ||||||||||
| Enterococcocal surface protein | – | – | – | – | – | – | – | – | – | – |
| Hystidine decarboxilase (2 sets of primers) | – | – | – | – | – | – | – | – | – | – |
| Hyaluronidase | – | – | + | – | – | – | – | – | – | + |
| Aggregation substance | – | – | – | – | – | – | – | – | – | – |
| Cytolisin | – | – | – | – | – | – | – | – | – | – |
| Endocarditis antigen | – | – | – | – | – | – | – | – | – | – |
| Tyrosine decarboxylase | + | + | + | + | + | + | + | + | + | + |
| Adhesion of collagen protein | – | – | – | – | – | – | – | – | – | – |
| Ornithine decarboxilase | + | + | + | + | + | + | – | + | + | + |
| Lb. plantarum S18 | Lb. plantarum S20 | Lb. plantarum S23 | Lb. brevis S27 | Lb. plantarum S29 | Lb. plantarum S30 | Lb. plantarum S35 | Lb. brevis S36 | Lb. plantarum S37 | Lb. brevis S38 | Lb. plantarum S39 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Bacteriocin genes | |||||||||||
| Curvacin A | – | – | – | – | – | – | – | – | – | – | – |
| Nisin | + | – | – | – | – | – | – | – | – | – | – |
| Pediocin PA1 | – | – | – | – | – | – | – | – | – | – | – |
| Plantaricin W | – | – | – | – | – | – | – | – | – | – | – |
| Plantaricin NC8 | – | – | – | – | – | – | – | – | – | – | – |
| Plantaricin S | – | – | – | – | – | – | – | – | – | – | – |
| Sakacin G | – | – | – | – | – | – | – | – | – | – | – |
| Sakacin T α | – | – | – | – | – | – | – | – | – | – | – |
| Sakacin T β | + | + | + | + | + | + | + | + | + | + | + |
| Sakacin X | + | – | + | + | – | – | + | + | + | – | – |
| Genes related to virulence factors, production of biogenic amines and vancomycin resistance | |||||||||||
| Vancomicin B | – | – | – | – | + | – | – | – | – | – | – |
| Vancomicin A | – | – | – | – | – | – | – | – | – | – | – |
| Gelatinase | + | ||||||||||
| Enterococcocal surface protein | – | – | – | – | – | – | – | – | – | – | – |
| Hystidine decarboxilase (2 sets of primers) | – | – | – | – | – | – | – | – | – | – | – |
| Hyaluronidase | – | – | – | – | – | – | – | – | – | – | – |
| Aggregation substance | – | – | – | – | – | – | – | – | – | – | – |
| Cytolisin | – | – | – | – | – | – | – | – | – | – | – |
| Endocarditis antigen | – | – | – | – | – | – | – | – | – | – | – |
| Tyrosine decarboxylase | + | + | + | + | + | + | + | + | + | + | + |
| Adhesion of collagen protein | + | – | – | – | + | – | – | – | – | – | – |
| Ornithine decarboxilase | + | + | + | – | + | – | – | – | – | – | + |
The selected microorganisms were tested for the harbouring of virulence genes gelE (gelatinase), hyl (hyaluronidase), asa1 (aggregation substance), esp (enterococcal surface protein), cylA (cytolisin), efaA (endocarditis antigen), ace (adhesion of collagen), vanA and vanB (both related to vancomycin resistance), and genes for amino acid decarboxylases: hdc1 and hdc2 (both related to histidine decarboxylase), tdc (tyrosine decarboxylase), and odc (ornithine decarboxylase) (Table 3), using the PCR protocols of Martin-Platero et al.20, Vankerckhoven et al.21 and Rivas et al.22. The amplified products were separated by electrophoresis on 0.8–2% (w/v) agarose gels in 0.5× TAE buffer. Gels were stained in 0.5× TAE buffer containing 0.5μg/ml ethidium bromide.
Results and discussionAcidification ability, growth at different temperatures, pH and NaCl levelsThe ability of LAB to acidify is one of the most important technological characteristics for potential starter cultures in the meat industry. The lowering of the pH of the LAB can inhibit the growth of a large number of pathogenic or undesirable microorganisms that cause spoilage of fermented meat products, and can improve the hygienic properties and storage of the final products. On the other hand, a pH of 5.1–5.3 has a positive effect on the humidity of the fermented meat products and accelerates the maturation process. The obtained results show that all tested strains, except for Lb. plantarum S23 and Lb. brevis S36, have good acid formation ability. The range of the acidifying ability for the studied strains was found to be from pH 4.03 to pH 5.2. For a correct fermentation process in the product pH must reach values near to the isoelectric point of fibrilar proteins. Proteolytic reactions that occur during fermentation of raw sausages have been studied extensively. Primarily, meat proteins are degraded into peptides by endogenous enzymes during fermentation of meat products. Since most bacteria (e.g., lactobacilli) grown in fermented meat products have only weak proteolytic activity, the degradation of proteins is not greatly affected by bacteria. However, LAB influence protein degradation by causing a decrease in pH, which results in increased activity of muscle proteases. More importantly, the peptides generated by muscle proteolysis can be taken up by bacteria that further split them intracellularly into amino acids and may convert them to aroma components.
The pH value and acidity are typical indicators of fermented meat products.23 Before drying, most dry sausages have similar acid activity. In some spontaneous fermentation processes pH values from 5.1 to 5.5 and a concentration of lactic acid of 0.3% to 0.9% have been reported.24 Natural LAB microbiota, particularly well adapted to these dry sausages, is characterized by a high acidifying potential. For the proper functioning of fermentation, the pH at the time of filling should reach 4.6–5.1, a value close to the isoelectric point of fibrillar proteins.31 Positive technological aspects of acidification, besides the inhibition of pathogenic microorganisms and faster drying of the product, are the improved texture through the denaturation and coagulation of proteins, the activation of muscle proteases and the formation of the red colour of the sausage by stimulating the formation of nitrite and nitrozomioglobulin.25
The ability of studied strains to develop in the medium at different initial pH (pH 3.0–pH 5.0) was monitored during their cultivation in MRS medium, along with their growth capability on acetate agar. The large majority of the strains isolated from the dry lukanka sausage were able to grow on acetate agar. From the studied strains only Lb. plantarum S7 and Lb. brevis S8 have good growth under acidic conditions (pH 3.3) (data not shown). For most (Lb. plantarum S7, S18 and S29, Lb. brevis S8, S36 and S38) pH 4.0 was more favourable. All strains grew very well in the neutral and weakly alkaline region, and with the exception of strains Lb. plantarum S23 and Lb. brevis S38, have very low growth in high alkaline conditions (pH 9.6). Some of the strains were characterized by a very narrow pH range of growth (Lb. plantarum S2, S3 and S7 grow well only at an initial pH of 7.0–7.5). Lb. plantarum S7 have a wide pH range, growing in the range of 3.3–8.5). Lb. brevis S38 has the ability to grow in the entire pH range investigated (pH 4.0–9.6). Many of the strains investigated (with the exception of Lb. plantarum S3, S18 and S37) were not able to grow at pH 8.5 and 9.6.
The ability of the cultures to grow in a particular temperature range is an important physiological characteristic used for the identification of LAB. For this purpose, isolates grown at temperatures of 15°C, 25°C, 37°C, 40°C, and 50°C were studied. Almost all strains tested had good (copious) growth at 37°C (with the exception of Lb. plantarum S3 and Lb. brevis S36), moderate growth at 40°C (with the exception of the strains Lb. plantarum S2 and S3 and Lb. brevis S36), and weak growth at 50°C (excluding Lb. plantarum S3). The growth of Lb. plantarum S37 was abundant at high temperatures. At 25°C most strains have abundant growth. Approximately half of the strains did not grow at 15°C. Lb. plantarum S29 and S39 had very good growth characteristics over a wide temperature range.
The resistance of LAB to the concentration of NaCl in the middle varies and is an important characteristic of their species differentiation. Furthermore, stability and growth in increasing concentrations of salt are important technological characteristics of the LAB of fermented meat(s). In this study, the ability of the LAB to grow has been examined in the growth medium supplemented with 2%, 4%, 6.5%, 10%, and 18% NaCl. The growth of most strains was abundant in the concentration of NaCl in the range of 2–6.5%. At 10% concentration of NaCl, many strains had moderate and abundant growth, but some (Lb. brevis S27 and Lb. plantarum S29) grew poorly, and only Lb. brevis S36 did not develop. In the presence of 18% NaCl all strains presented poor growth.
Technological properties of sodium chloride are related to its effect on the flavour, texture and shelf life of meat products. In search of healthier fermented food products, in most cases a salt content of over 2% can be markedly lowered without substantial sensory deterioration or technological problems leading to economical losses. Sodium chloride content down to 1.4% in cooked sausages and 1.75% in lean meat products is considered to be enough to produce a heat-stable gel with acceptable perceived saltiness as well as firmness, water-binding and fat retention. However, a particular problem with low-salt meat products is that not only the perceived saltiness, but also the intensity of the characteristic flavour can be affected. Increased meat protein content (i.e. lean meat content) in meat products reduces the perceived saltiness. From the technological point of view, the required salt content for acceptable gel strength depends on the formulation of the product. However, when phosphates are added or the fat content is high, lower salt additions provide a more stable gel than in non-phosphate and in low-fat products. Small differences in salt content at the 2% level do not have a marked effect on the shelf life of the products. By using salt mixtures, usually NaCl/KCl, the intake of sodium (NaCl) can be further reduced.11
Proteolytic activityThe strains cultured in milk and analyzed by SDS-PAGE, showed different profiles related to the proteolytic activity. The performed analysis determined that the tested strains hydrolysed high molecular weight proteins of the milk with varying capacity. The residue amounts of k-casein and α, β-casein were below 50% within the majority of the strains. Low molecular weight proteins of the milk were also hydrolysed by most of the studied strains. It should be noted that some strains hydrolysed β-lactoglobulin completely. According to previously reported data, the LAB have a relatively low proteolytic activity against myofibrillar proteins.23 However, some strains of Lb. casei, Lb. plantarum, Lb. curvatus, and Lb. sakei contributed significantly to the hydrolysis of sarcoplasmic proteins, as well as to the subsequent decomposition of the peptides to free amino acids.23 Furthermore, some strains of Lb. sakei, Lb. curvatus, and Lb. plantarum showed the presence of leucine and valine amino peptidases, which contribute to the breakdown of proteins and peptides. In this context, amino acid release, attributed to precursors, is responsible for the flavour of the final product.11 All these results indicate that milk proteins may be used as a model system to establish the proteolytic activity of strains. The analysis of proteolysis in the manufacture of fermented sausages, screening for proteinases, peptidases, and aminopeptidases during the selection of starters should be recommended as part of the screening for new starter cultures with beneficial properties.
Enzyme activitiesIn Table 2 the activities of the enzymes correlated with carbohydrate catabolism are shown and they differ considerably. β-Galactosidase activity was exhibited by one strain of Lb. plantarum (S3) and three strains of Lb. brevis (S8, S27 and S38). In contrast, the majority of the strains exhibit α-galactosidase activity (24 strains). Almost all strains have β-glucosidase activity (except for Lb. sakei S13 and S15), while the number of strains with α-glucosidase activity was considerably lower. The results for α-galactosidase, β-glucuronidase, α-mannosidase, and N-acetyl-β-glucosaminidase activity, found in the major proportion of strains, confirmed those obtained from Papamanoli et al.11. The acid taste of fermented meat products, which is correlated to the acid content, is one of the components of the overall taste.26 Metabolism during fermented dry sausage ripening is a complex interaction between residual enzyme activity of the muscle and/or fat tissue and bacterial metabolism.
With respect to lipolytic activity for the strains isolated from lukanka, our studies have shown that all strains have a very low activity. These data are consistent with the referred to in the work by Montel et al.26 According to Vestergaard et al.,27 the lipolysis of meat products is mainly due to muscle lipase and phospholipase, while bacterial lipases show very little activity in terms of fermented products.28
The degradation of amino acids to volatile molecules played an important role in determining the characteristic taste of dry sausages. Studies by Montel et al.26 showed that the aldehydes, alcohols and acids derived from the breakdown of leucine, valine, phenylalanine and methionine have very low values. The examined activity of valine arylamidase and cysteine arylamidase detected when using APIZYM for the majority of strains in our study was high (Table 2). The activity of leucinearylamidase is relatively low for most strains, while for some strains of Lb. plantarum (S2, S5, and S7) and L. sakei (S11) was not detected. Acid phosphatase and alkaline phosphatase activities were not exhibited by most of the tested strains, but all four Lb. brevis have good alkaline activity.
Spectrum of antimicrobial activityThe studied LAB were evaluated for their antimicrobial activity using the agar-spot-test against a panel of test organisms (Table 1). Cell-free supernatants obtained from Lb. plantarum (BS7, BS9, BS20, BS30, BS37 and BS39) and Lb. brevis (BS8, BS36) (adjusted to pH 6.0–6.5) inhibited the growth of several strains belonging to L. monocytogenes. However, none of the other bacteria included in the microorganism panel test was affected (Table 1). This narrow spectrum of bacteriocin activity was revealed as being unique for Lactobacillus spp. In fact, most of the bacteriocins described for Lactobacillus spp. were found to be active against a much broader range of microbial genera and species.19 However, it is important to emphasize that the very strong activity against L. monocytogenes exhibited by BS7, BS30, BS36, BS37 and BS39 strains could have significant application in the biopreservation of fermented food products. However, to be not active against various LAB and to inhibit various L. monocytogenes suggests a very good potential for this bacteriocin(s) or appropriate LAB to be used in the biopreservation of various food products while at the same time not affecting the applied starter cultures.
Screening for presence of bacteriocin genesOn the basis of the PCR reactions performed targeting nisin, pediocin PA-1, curvacin A, plantaricin S, plantaricin NC8, plantaricin W, sakacin G, sakacin Tα, sakacin Tβ and sakacin X, some of the tested strains generated positive results (Table 3). The sequence of the generated amplicons was 100% identical to the targeted bacteriocin genes. All the strains produced positive results for the presence of sakacin Tβ, however none of them generated evidences for the presence of sakacin Tα. It is important to refer to the fact that sakacin Tα and sakacin Tβ are part of the two-bacteriocin complex, and the presence of both peptide is required for the activity of a mature peptide system.17 Carrying only one of the required genes for two-peptide bacteriocins is most probably related to the fact that primary solicitation of sakacin Tβ in the life circle of the tested strains is not an antimicrobial and defence protein, but probably has a different role. Recently raised the hypothesis that some of the polypeptides produced by the LAB has a regulatory function, but because of the shared similarity to some antimicrobial peptides they exhibit some antimicrobial activity as well.29,30
We need to point out another fact: some of the tested LAB has generated antibacterial activity based on the agar-spot-test method, but we cannot detect the presence of the tested bacteriocin genes (Tables 1 and 3) and vice versa. It has been previously reported that specific LAB strains can carry more than one bacteriocin gene, but cannot express some of them.31 However, additional experiments on bacteriocin purification, and the amino-acid sequence of the expressed bacteriocin(s) is required in order to confirm the expression of the detected bacteriocin gene(s). From this perspective, it seems important to point out that the application of different approaches for the characterization of a bacteriocin is crucial. Very frequently authors report on new bacteriocin identification merely based on the determination of the presence of gene(s) for bacteriocin(s) production.32,33 Albano et al.34 verified the presence of pediocin PA-1 genes in two different strains of P. acidilactici isolated from “Alheira.” However, the evidences for the expression of these bacteriocins were not reported and the studied bacteriocins have been only presumed to be expressed.32–34 The purification of the bacteriocin followed by mass spectrometry and partial or complete amino acid sequencing is proof that the genes are actually being expressed.35 Moreover, Poeta et al. showed that some bacteriocin-producer strains may carry several genes for bacteriocin production and, depending on growth conditions, they could express one or another gene.31 As a consequence, reports based only on the detection or identification of bacteriocin genes need to be considered with high scepticism and results should be accepted only when additional research and sufficient evidence of bacteriocin production and expression is made available.
Screening for physiological and genetic trait related to virulence factor, biogenic amines and antibiotic resistanceDespite being very relevant in food isolates and well accepted as GRAS microorganisms, the verification of virulence factors in Lactobacillus spp. by bio-molecular procedures is necessary due to the risk of genetic transfer, since these genes are usually located in conjugative plasmids.36 Interestingly, from all the screened genetic sequences related to several virulence factors, biogenic amines and antibiotic resistance (Table 3), only PCR targeting vancomycin B (in 3 from 21 tested isolates), gelatinase (in 1 from 21 tested isolates), hyaluronidase (in 2 from 21 tested isolates), tyrosine decarboxylase (in 21 from 21 tested isolates), ornithine decarboxilase (in 2 from 21 tested isolates) and adhesion of collagen protein (in 2 from 21 tested isolates) genes generated positive results. VanB genes confer resistance to various concentrations of vancomycin only and have been described in various LAB.37
The presence of the virulence factors is an important point in the safety evaluation of LAB with a potential application as functional strains (probiotic or starter cultures). Even if these virulence factors have been described as playing a role in the pathogenicity of the Enterococcus spp., we need to consider their presence in functional LAB with high precision. The scenario of horizontal gene transfer is a very high possibility in the mixed microbial population and we need to be aware of this fact.
ConclusionsFactors such as the physiological state of LAB as starters, the physical conditions of ripening and storage (e.g. temperature, pH, sodium chloride), chemical composition of the matrix (acidity, available carbohydrate content, water activity, etc.) and the possible interaction of the starter cultures with probiotic and/or other microorganisms occurring naturally or added to the system of the studied meat product have to be considered when develop new functional fermented food products.
Some metabolites such as antimicrobial peptides can play a role in LAB performances and metabolism, affecting not only microbial safety, but also the total population and the ecology of fermented meat products. Very strong activity against L. monocytogenes was detected for some of the studied LAB. In addition, the studied strains did not present any antimicrobial activity against tested closely related bacteria, and frequently used as starter cultures, such as Lactobacillus spp., Lactococcus spp., Enterococcus spp. or Pediococcus spp. Studied LAB presented low levels of presence of virulence factors, including antibiotic resistance and biogenic amines related genes and may be considered as safe, in addition to positive technological characteristics for production of fermented food products such as acidification properties, survival at increased NaCl concentrations, production of technologically important enzymes and proteolitic activity.
Conflicts of interestThe authors declare no conflicts of interest.
The authors would like to acknowledge CNPq 313852/2013-8 and CNPq 401140/2014-8 projects for providing financial support in addition to support from CAPES and FAPEMIG.