High potential, thermotolerant, ethanol-producing yeasts were successfully isolated in this study. Based on molecular identification and phylogenetic analysis, the isolated thermotolerant yeasts were clustered in the genera of Pichia kudriavzevii, Candida tropicalis, Candida orthopsilosis, Candida glabrata and Kodamea ohmeri. A comparative study of ethanol production using 160g/L glucose as a substrate revealed several yeast strains that could produce high ethanol concentrations at high temperatures. When sugarcane bagasse (SCB) hydrolysate containing 85g/L glucose was used as a substrate, the yeast strain designated P. kudriavzevii RZ8-1 exhibited the highest ethanol concentrations of 35.51g/L and 33.84g/L at 37°C and 40°C, respectively. It also exhibited multi-stress tolerance, such as heat, ethanol and acetic acid tolerance. During ethanol fermentation at high temperature (42°C), genes encoding heat shock proteins (ssq1 and hsp90), alcohol dehydrogenases (adh1, adh2, adh3 and adh4) and glyceraldehyde-3-phosphate dehydrogenase (tdh2) were up-regulated, suggesting that these genes might play a crucial role in the thermotolerance ability of P. kudriavzevii RZ8-1 under heat stress. These findings suggest that the growth and ethanol fermentation activities of this organism under heat stress were restricted to the expression of genes involved not only in heat shock response but also in the ethanol production pathway.
Ethanol production is not economically feasible in tropical countries because of the high energy requirements for cooling systems. To reduce the operating costs and to improve ethanol production efficiency, the development of a new technology is necessary. High-temperature ethanol fermentation (HTEF) is of great interest because it has several advantages, e.g., reduced cooling costs, reduced contamination risk by undesirable mesophilic microorganisms, and increased rates of sugar to ethanol conversion, resulting in an increase in ethanol productivity.1,2 Many researchers have attempted to explore and characterize effective, thermotolerant, ethanol-producing yeasts that are capable of growth and ethanol production at high temperatures. There are several yeast species that have been characterized and classified as thermotolerant yeasts, such as Kluyveromyces marxianus, Pichia sp., Candida sp., and some strains of Saccharomyces cerevisiae.2–5 Recently, there are several reports on ethanol production at high temperatures using Pichia kudriavzevii (formally known as Issatchenkia orientalis).6–10 Several strains of P. kudriavzevii have been reported to grow and produce high levels of ethanol at high temperatures, e.g., P. kudriavzevii MF-12111 and P. kudriavzevii DMKU 3-ET15.12 In addition, some strains of P. kudriavzevii, such as strain MF-121, also exhibit multi-stress tolerance, such as acid, ethanol, thermo, and salt tolerance.13,14S. cerevisiae, the yeast traditionally used for ethanol production, shows high ethanol fermentation efficiency and high ethanol tolerance. In comparison, P. kudriavzevii exhibits higher thermotolerance ability than S. cerevisiae.15,16P. kudriavzevii has been isolated from many sources, e.g., pork sausage,12 cheese,17 grape wine pomace,18 cocoa beans,19 and rice bran,20 but to the best of our knowledge, there are no reports on the isolation of this yeast strain from plant orchards.
There are several feedstocks that can be used to produce bioethanol on an industrial scale. Among these feedstocks, lignocellulosic biomass is considered an attractive feedstock to produce fuel ethanol because it is available in large quantities at a low cost, and it is a very promising feedstock for reducing competition with food.21,22 One of the major types of lignocellulosic biomass that is found in great quantities at a low cost, particularly in Thailand, is sugarcane bagasse (SCB). In the sugar production process, 1 ton of sugarcane generates approximately 280kg of bagasse.23 SCB is composed of 40–45% cellulose, 30–35% hemicellulose and 20–30% lignin.24 Due to its lower ash content (1.9%), it offers several advantages compared with other agricultural residues, such as paddy straw, rice straw and wheat straw.25–27 In terms of its potential, SCB is attracting attention as a highly promising raw material for industrial ethanol production.
Heat stress during ethanol fermentation has been reported to stimulate the expression of several genes encoding heat shock proteins (HSPs) and proteins involved in the ethanol biosynthesis pathway, trehalose and glycogen metabolism, ATP production and the protein degradation processes of K. marxianus and S. cerevisiae.2,5,28 However, less is known about the heat-shock response in thermotolerant P. kudriavzevii. In general, HSPs play an important role in conferring thermotolerance by protecting polypeptides from being irrevocably denatured, misfolded or aggregated under various exogenetic and abiotic stress conditions. They also help organisms modulate their stress responses and protect themselves from cellular damage.29,30 In addition, some molecules, such as trehalose and glycogen, can also confer thermotolerance to microorganisms.31 Trehalose has been reported to protect yeast cells by stabilizing proteins and membranes under stress conditions, especially during desiccation and heat stress.32 Mahmud et al.33 reported that high trehalose accumulation could make the yeast cells resistant to multiple stresses, such as ethanol, heat, and freezing; however, the importance of this accumulation before and after stress induction varied, depending on the type of stress. Glycogen has also been reported to be involved with tolerance toward several stresses, such as heat, osmotic and oxidative stress.2,34 While many genes responsible for stress tolerance in yeast cells have been reported, it is still unclear whether the expression of genes during stress conditions is directly related to stress tolerance, especially in P. kudriavzevii. In this study, we investigated the isolation and characterization of thermotolerant yeasts collected from plant orchard samples that were capable of growth and ethanol production at high temperatures. Molecular identification and ethanol fermentation efficiency tests of the selected thermotolerant yeasts were also performed. Furthermore, to gain a better understanding of the molecular mechanisms involved in the acquisition of thermotolerance during HTEF in the selected thermotolerant yeast, the expression of genes encoding HSPs and proteins involved in the pyruvate-to-ethanol (PE) pathway, fructose 1,6-bisphosphate aldolase-to-pyruvate (FP) pathway, and trehalose and glycogen metabolism was determined using quantitative real-time polymerase chain reaction (qRT-PCR). (A part of this work has been presented as a proceeding in the 26th Annual Meeting of the Thai Society for Biotechnology and International Conference; TSB 2014).
Materials and methodsIsolation, screening and selection of thermotolerant ethanol-producing yeastsSoil, plant bark decay, manure and rotten fruits collected from various plant orchards in the Chon Buri, Rayong, Chanthaburi and Khon Kaen provinces, Thailand, were subjected to the isolation of thermotolerant yeasts using a YM medium supplemented with 4% (v/v) ethanol and incubated at 35°C. All procedures for the isolation of thermotolerant yeasts were performed according to the method described by Limtong et al.3 All pure cultures were collected and maintained on YM agar at 4°C for short-term storage and in 50% (v/v) glycerol at −20°C for long-term storage.
Screening of the thermotolerant ethanol-producing yeasts was performed by culturing each isolate of the yeast in a 20×20mm test tube containing 15mL of YM liquid medium and then incubating the cultures at 37°C for 48h on a rotary incubator shaker at 150rpm. A Durham tube was placed inside the test tube, and the yeast strains that could ferment glucose and accumulate a high level of CO2 in the Durham tube were selected for further experiments. All experiments were carried out in duplicate and repeated twice.
Identification of the selected thermotolerant yeastsThe selected thermotolerant yeasts were identified by molecular analysis based on the D1/D2 domain of the 26S rDNA gene sequence.35 Genomic DNA was isolated from the selected thermotolerant yeasts using the method described by Harju et al.,36 and it was then subjected to polymerase chain reaction (PCR) using the primers NL-1 and NL-4,37 which were specific for the D1/D2 region of the large subunit of the 26S rDNA gene. The reaction mixtures (50μL) and the PCR amplification conditions were performed using the Taq PCR core kit (QIAGEN, Germany). All procedures were carried out as described in the manufacturer's instructions. After PCR amplification, the PCR product was separated using a 1% agarose gel and purified using an Invisorb® Fragment CleanUp kit (Invitek GmbH, Berlin, Germany). The purified PCR product was subjected to DNA sequencing (the First BASE Laboratories Sdn Bhd, Seri Kembangan, Selangor Durul Ehsan, Malaysia) and was analyzed using GENETYX (Software Development, Tokyo, Japan). Homology and phylogenetic analyses were performed using the FASTA and BLAST programs from the GenBank database and MEGA5,38 respectively. Tree topologies were constructed based on the neighbor-joining method with 1000 bootstrap replicates.39
Ethanol production at high temperatures by the selected thermotolerant yeasts using glucoseThe ethanol production capability of the selected thermotolerant yeasts was evaluated at high temperatures using a YM medium containing 16% (w/v) glucose.40 The active yeast cells (1×107cells/mL) were inoculated into 100mL of YM medium and then incubated on a rotary incubator shaker (150rpm) at the high temperatures of 37, 40 and 45°C. During ethanol fermentation, fermentation broth was taken at certain time intervals, and the ethanol concentration was measured using gas chromatography (GC).
Ethanol production at high temperatures by the selected thermotolerant yeasts using SCB hydrolysateSCB was used as a substrate for high-temperature ethanol production by the selected thermotolerant yeasts. SCB was collected from Mitr Phuvieng Sugar Factory, Khon Kaen province, Thailand. Preparation of the SCB hydrolysate was performed using the method described by Srinorakutara et al.41 Cellic® CTec2 (Novozymes, Denmark) at a concentration of 50FPU/g DS was added to 15g of acid-pretreated SCB, then incubated at 50°C on a rotary shaker at 160rpm for 48h. The slurry obtained from enzymatic hydrolysis was centrifuged at 8000rpm for 10min, and the supernatant (SCB hydrolysate) was used for the ethanol fermentation experiments. The glucose concentration in the SCB hydrolysate was measured using high-performance liquid chromatography (HPLC).
Batch ethanol fermentation was performed in a 500mL Erlenmeyer flask containing 200mL of SCB hydrolysate supplemented with 2g/L yeast extract, 2g/L peptone and 2g/L MgSO4.42 The active yeast cells (1×107cells/mL) were inoculated into the flask and then incubated at the high temperatures of 37, 40 and 45°C in a rotary incubator shaker at 150rpm. During ethanol fermentation, fermentation broth was randomly collected at certain time intervals. Sugar and ethanol concentrations in the fermentation broth were analyzed using the phenol–sulfuric acid method43 and GC, respectively.
Growth characterization of the thermotolerant yeast P. kudriavzevii RZ8-1 under different stress conditionsGrowth of the isolated thermotolerant yeast P. kudriavzevii RZ8-1 under different stress conditions, including heat, ethanol and acetic acid stress, was determined using the method described by Yuangsaard et al.,12 with slight modifications. Yeast cells were cultured in a YM broth at 35°C in a rotary incubator shaker at 150rpm. After 24h of incubation, the culture broth was transferred to a fresh YM medium and subsequently incubated under the same conditions until the exponential growth phase was reached. Cells were collected by centrifugation at 5000rpm and subjected to a serial dilution; then, 10μL of the appropriated dilution culture was spotted onto a culture medium. For heat stress treatment, cells were cultured on YM agar and incubated at the high temperatures of 37, 40, 42 and 45°C. For ethanol stress treatment, cells were cultured on YM agar containing different ethanol concentrations of 4, 6, 8, 10, 12 and 16% (v/v) and incubated at 35°C. For acetic acid stress treatment, cells were cultured on YM agar containing acetic acid at concentrations of 0.5, 1.0, 2.5, 5.0 and 7.5g/L and then incubated at 35°C. After 24h of cultivation, growth of the thermotolerant yeast P. kudriavzevii RZ8-1 on agar medium was monitored.
Viability test of P. kudriavzevii RZ8-1 during fermentation at high temperatureThe viability of the thermotolerant yeast P. kudriavzevii RZ8-1 during fermentation at high temperature (40°C) was evaluated using repeated batch fermentation. It was carried out in a 500-mL Erlenmeyer flask containing 200mL of SCB hydrolysate supplemented with 2g/L yeast extract, 2g/L peptone and 2g/L MgSO4.42 The active yeast cells (1×107cells/mL) were inoculated into the flask and then incubated at 40°C in a rotary incubator shaker at 150rpm for 24h. Thereafter, the fermentation broth was withdrawn at the end of each batch, and the number of viable and dead cells were analyzed. Fresh fermentation medium was added to the flasks for subsequent batch fermentation.
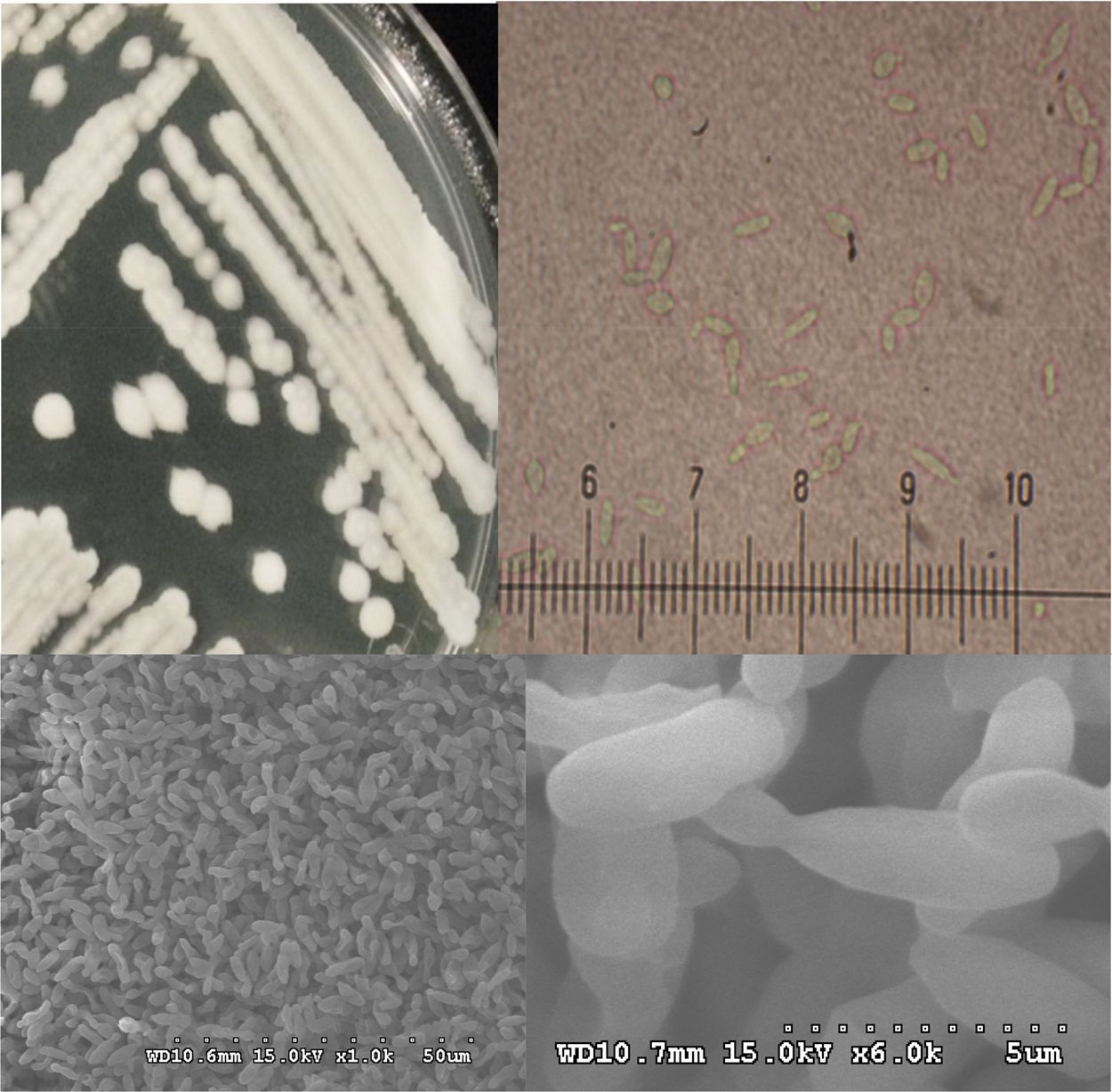
Morphological characterization of the thermotolerant yeast P. kudriavzevii RZ8-1Cell morphology of the thermotolerant yeast P. kudriavzevii RZ8-1 was determined using bright field microscopy (Carl Zeiss Primo star microscope, Carl Zeiss Microscopy, Germany) and scanning electron microscopy (SEM; Hitachi model S-3000N, Tokyo, Japan). A single colony of yeast cells was cultured on YM agar and incubated at 35°C for 24h. Morphology of the colony was monitored using a microscope and was photographed. For SEM analysis, cells were grown in YM broth at 35°C in a rotary incubator shaker at 150rpm. After 24h of incubation, cells were collected by centrifugation, washed twice with sterile distilled water and then frozen in a vacuum freeze-drying machine (CHAIST, Germany). The resulting cells were sputtered with gold (EMITECH model K500X, Kent, UK) and then photographed.
Ethanol fermentation and gene expression analysis using quantitative real time-PCR (qRT-PCR)The batch ethanol fermentation by the selected thermotolerant yeast P. kudriavzevii RZ8-1 was carried out using a YM medium containing 100g/L glucose. The active yeast cells (1×107cells/mL) were inoculated into a YM medium and incubated on a rotary incubator shaker (150rpm) at 30 and 42°C. Samples were withdraw at certain time intervals during ethanol fermentation and the ethanol concentration and the number of viable cells were analyzed.
The expression of genes encoding HSPs and the proteins involved in the glycolysis pathway and trehalose and glycogen metabolism was evaluated during ethanol fermentation at high temperature (42°C) using qRT-PCR. For normal conditions (control experiment), the active yeast cells (1×107cells/mL) of the selected thermotolerant yeast P. kudriavzevii RZ8-1 were inoculated into a YM medium containing 100g/L glucose and incubated in a rotary incubator shaker (150rpm) at 30°C for 9h (exponential growth phase). For heat shock conditions, cells that were in the exponential growth phase in a YM medium containing 100g/L glucose at 30°C were shifted to 42°C and subsequently incubated in a rotary incubator shaker at 150rpm for 2h. For long-term heat stress conditions, the active yeast cells were cultured in a YM medium containing 100g/L glucose and incubated in a rotary incubator shaker (150rpm) at 42°C for 9h. After incubation, cells were collected by centrifugation at 5000rpm for 10min, washed twice with sterile distilled water, and then subjected to RNA isolation using TRIzol® reagent (Thermo Fisher Scientific, Waltham, MA). All procedures for RNA isolation were performed according to the manufacturer's instructions. The RNA concentration was measured and adjusted by Nanodrop (Nanodrop Technologies, Wilmington, DE, USA). qRT-PCR amplification was carried out using a Biorad-I-Cycler with qPCRBIO SyGreen One-Step PCR-Biosystems (PCRBIOSYSTEMS, London, UK). The reaction mixtures (20μL) and the thermal cycling for qRT-PCR amplification were carried out according to the manufacturer's instructions. The primers used for qRT-PCR amplification are listed in Table 1. Based on the agarose gel electrophoresis analysis of the qRT-PCR products, only a single band of DNA was observed, and the size of the amplicons was varied, ranging from 177 to 587bps, depended on the genes. The actin gene (act) and RNase-free water were used as internal and negative controls, respectively. The fold change of transcript levels was calculated using the 2−ΔΔCt method.44
Primers used for quantitative real-time polymerase chain reaction (qRT-PCR) analysis in this study.
| No. | Primer name | Gene name | Sequence (5′–3′) | PCR product (bp) |
|---|---|---|---|---|
| 1 | HSP90-F | Heat shock protein 90 | TCTACCAAGCTTTCTCTAAG | 177 |
| HSP90-R | CGGTAATGAAGTAGATGTTC | |||
| 2 | Ssq1-F | Heat shock protein 70 family | GAGAATCACGAGGGACAAAG | 558 |
| Ssq1-F | GAGAATCACGAGGGACAAAG | |||
| 3 | NTH1 606-F1 | Neutral trehalase | GGCCAAAAAGTGATCCTTCA | 572 |
| NTH1 606-R1 | GGTTTTTCGCCTTTGGTGTA | |||
| 4 | NTH1572-F2 | Neutral trehalase | TAGATCACAGCCACCCTTCC | 546 |
| NTH1572-R2 | TTCAGCCAACTTCTCCCAGT | |||
| 5 | GGS1-F | Trehalose synthase | CCTTGAACGAATGTCCTGCT | 537 |
| GGS1-R | GGGAACGGCGTATGTAAGAA | |||
| 6 | ADH1-F | Alcohol dehydrogenase 1 | ACGTTAAGTACTCTGGTGTG | 578 |
| ADH1-R | CATCCAATACGCCAAGGCTA | |||
| 7 | ADH2-F | Alcohol dehydrogenase 2 | CGACGGTTCTTTCCAACAAT | 524 |
| ADH2-R | ATCGTTGGTTCTTACGTCGG | |||
| 8 | ADH3-F | Alcohol dehydrogenase 3 | GGCCTCTGTTGCTATTCCAG | 565 |
| ADH3-R | TGGTGGTCTAGGTTCCTTGG | |||
| 9 | ADH4-F | Alcohol dehydrogenase 4 | GGCTCCAGAAGACTATTCGT | 525 |
| ADH4-R | CACCGTCTACAAGGCTCTAA | |||
| 10 | GSK3-F | Glycogen synthase kinase | CAACCCGAGGAGTCAGTTGT | 505 |
| GSK3-R | ATGACACATTGGGCTCGTTT | |||
| 11 | TDH2-F | Glyceraldehyde-3-phosphate dehydrogenase | TTTCCAACGCTTCCTGTACC | 540 |
| TDH2-R | CGTCAAGTTGGTTTCCTGGT | |||
| 12 | ENO-F | Enolase | GAACGTCTTGAACGGTGGTT | 535 |
| ENO-R | TGTCACCAACCCAGTCAGAA | |||
| 13 | Actin-F | Actin | CATTCAAGCCGTTTTGTCCT | 559 |
| Actin-R | GGAAATCACTGCTTTGGCTC |
The glucose concentration was measured by HPLC using an Aminex HPX87H column (300×7.8mm) (Bio-Rad, Hercules, CA, USA) with a refractive index detector at 50°C. Sulfuric acid (5mM) was used as a mobile phase at a flow rate of 0.6mL/min. The ethanol concentration was determined by GC (Shimadzu GC-14B, Japan) using a polyethylene glycol (PEG-20 M) pack column with a flame ionization detector (FID). Isopropanol was used as an internal standard.45 The volumetric ethanol productivities (Qp, g/Lh), ethanol yields (Yp/s, g/g) and theoretical ethanol yields (T.Y, %) were calculated using the method described by Nuanpeng et al.2 The yeast cell number was determined using a hemocytometer, and cell viability was assessed by staining with 0.1% methylene blue.46
All experiments were conducted in duplicate and repeated twice, and the standard deviation (SD) values were calculated using MS Excel software. Multiple comparison tests for each experimental treatment were carried out by using Duncan's Multiple Range Test (DMRT) at the 5% probability level (p=0.05) in the SPSS program.
ResultsIsolation, screening and selection of thermotolerant ethanol-producing yeastsA total of 127 isolates of yeast were obtained after isolation using the enrichment culture technique described by Limtong et al.3 Most of the yeasts came from soil samples and rotten fruits. They were all tested for their ability to grow on YM agar at the high temperatures of 37, 40 and 45°C. As found in this study, 62 isolates of yeast could grow at 37°C, whereas 40 isolates could grow at 40 and 45°C (data not shown). Based on the definition of a thermotolerant yeast given by Sree et al.,47 40 isolates of yeast were classified as thermotolerant yeast since they could grow at temperatures higher than 40°C. To select for the thermotolerant fermentative yeast, all 40 isolates were cultured in 15mL of YM broth, and their abilities to ferment glucose and accumulate CO2 in the Durham tube were monitored. The results showed that all 40 isolates of the selected thermotolerant yeasts accumulated relatively high levels of CO2 in the Durham tube within 24h (data not shown). Therefore, they were subjected to identification using molecular techniques.
Identification of the selected thermotolerant yeastsAll 40 isolates of the selected thermotolerant yeasts were identified by the D1/D2 domain of the 26s rDNA gene sequence. Based on the phylogenetic analysis shown in Fig. 1, the yeasts were categorized into five groups. Nineteen isolates of yeast (KK07, CTB014, CTB011, CHOB1102, CTB15, CHOB03, KK14, CTB019, CHOB09, CTB013, CTB018, RZ10, RZ017, CHOB08, KK012, CTB016, CTB010, KK015 and RZ2305) were clustered into a group of Candida tropicalis, 15 isolates (RZ3-1, S10, S6, RZ06, RZ8-1, CHOB17, CTB18, KK19, KK21, CTB1-2, S11, CHOB1103, CHOB02, RZ5-1 and RZ11) were closely related to P. kudriavzevii, 4 isolates (KK09, CHOB01, CTB20 and KK22) were clustered into the same group as C. grabrata and C. albicans, 1 isolate (KK13) was closely related to C. orthopsilosis, and the remaining isolate (CTB16) was closely related to Kodamea ohmeri. These results suggested that a plant orchard was one of the best ecosystems for the isolation of thermotolerant fermentative yeast. It was also evident from these findings that almost all of the thermotolerant yeast that were isolated from the plant orchard belonged to the genus Candida, suggesting that this genus was predominant in this habitat.
Ethanol production at high temperatures by the selected thermotolerant yeasts using glucoseEthanol production capability at high temperatures (37, 40 and 45°C) by 40 isolates of the selected thermotolerant yeasts was determined using a YM medium containing 16% glucose, and the results are summarized in Table 2. At 37°C, the maximum ethanol concentrations and volumetric ethanol productivities produced by each isolate of yeast ranged from 27.49 to 64.87g/L and 0.53 to 1.35g/Lh, respectively. At 40°C, the highest ethanol concentrations and volumetric ethanol productivities ranged from 22.78 to 70.42g/L and 0.38 to 1.46g/Lh, respectively. At the highest temperature (45°C), the ethanol concentrations and volumetric ethanol productivities were remarkably lower compared with those at 37 and 40°C. Of the 40 isolates of the selected thermotolerant yeasts, 6 isolates of P. kudriavzevii (designated RZ3-1, RZ5-1, RZ8-1, S6, S10 and S11) exhibited relatively high levels of ethanol concentrations and volumetric ethanol productivities at 37, 40 and 45°C compared to the other tested isolates. Therefore, these 6 isolates of the selected thermotolerant yeasts were chosen for the next series of experiments.
Ethanol production at high temperature by the selected thermotolerant yeasts using glucose as a substrate.
| No. | Isolate | 37°C | 40°C | 45°C | |||
|---|---|---|---|---|---|---|---|
| P (g/L) | Qp (g/Lh) | P (g/L) | Qp (g/Lh) | P (g/L) | Qp (g/Lh) | ||
| 1 | CHOB01 | 48.27±0.08 (60h) | 0.80±0.00 | 45.65±0.03 (60h) | 0.76±0.00 | 28.20±0.04 (60h) | 0.47±0.00 |
| 2 | CHOB02 | 42.39±0.36 (60h) | 0.71±0.01 | 44.03±0.20 (60h) | 0.73±0.00 | 33.01±0.18 (60h) | 0.55±0.00 |
| 3 | CHOB03 | 37.56±0.69 (48h) | 0.78±0.01 | 30.70±0.28 (60h) | 0.51±0.00 | 6.01±0.26 (60h) | 0.10±0.00 |
| 4 | RZ3-1 | 60.46±0.30 (60h) | 1.01±0.00 | 67.86±0.17 (60h) | 1.13±0.00 | 43.75±0.12 (48h) | 0.91±0.00 |
| 5 | RZ5-1 | 61.88±0.19 (48h) | 1.29±0.00 | 70.51±0.10 (60h) | 1.18±0.00 | 45.10±0.10 (48h) | 0.94±0.00 |
| 6 | RZ06 | 59.84±0.50 (48h) | 1.25±0.01 | 62.18±0.21 (60h) | 1.04±0.00 | 27.99±0.26 (36h) | 0.78±0.01 |
| 7 | KK07 | 49.90±0.16 (60h) | 0.83±0.00 | 48.74±0.59 (60h) | 0.81±0.01 | 16.18±0.11 (36h) | 0.45±0.00 |
| 8 | RZ8-1 | 59.55±0.63 (48h) | 1.24±0.01 | 69.85±0.18 (48h) | 1.46±0.00 | 35.14±0.07 (36h) | 0.98±0.00 |
| 9 | KK09 | 41.96±0.16 (60h) | 0.70±0.00 | 45.008±0.13 (60h) | 0.75±0.00 | 35.70±0.22 (36h) | 0.99±0.01 |
| 10 | RZ10 | 43.97±0.31 (60h) | 0.73±0.01 | 52.56±0.82 (60h) | 0.88±0.01 | 14.07±0.42 (48h) | 0.29±0.01 |
| 11 | RZ11 | 60.59±0.57 (48h) | 1.26±0.01 | 40.81±0.14 (36h) | 1.13±0.00 | 32.76±0.24 (36h) | 0.91±0.01 |
| 12 | KK13 | 34.28±0.31 (60h) | 0.57±0.01 | 22.79±0.03 (60h) | 0.38±0.00 | 3.65±0.13 (60h) | 0.06±0.00 |
| 13 | KK14 | 49.03±0.02 (60h) | 0.82±0.00 | 23.01±0.01 (24h) | 0.96±0.00 | 13.35±0.18 (36h) | 0.37±0.00 |
| 14 | CTB15 | 56.58±0.00 (60h) | 0.94±0.00 | 24.62±0.20 (60h) | 0.41±0.00 | 13.22±0.06 (36h) | 0.37±0.00 |
| 15 | CTB16 | 36.60±0.01 (36h) | 1.02±0.00 | 35.87±0.10 (48h) | 0.75±0.00 | 4.14±0.27 (36h) | 0.11±0.01 |
| 16 | CHOB17 | 54.00±0.08 (48h) | 1.13±0.00 | 36.01±0.17 (36h) | 1.00±0.00 | 28.79±0.37 (48h) | 0.60±0.01 |
| 17 | CTB18 | 51.54±0.30 (48h) | 1.07±0.01 | 35.94±0.74 (36h) | 1.00±0.02 | 51.54±0.15 (48h) | 1.07±0.00 |
| 18 | KK19 | 53.93±0.29 (48h) | 1.12±0.01 | 35.49±0.07 (36h) | 0.99±0.00 | 52.37±0.74 (60h) | 0.87±0.01 |
| 19 | CTB20 | 32.13±0.15 (60h) | 0.54±0.00 | 31.79±0.65 (60h) | 0.53±0.01 | 28.38±0.09 (36h) | 0.79±0.00 |
| 20 | KK21 | 42.59±0.71 (36h) | 1.18±0.02 | 55.84±0.26 (48h) | 1.16±0.01 | 31.43±0.18 (24h) | 1.31±0.01 |
| 21 | KK22 | 27.49±0.22 (48h) | 0.57±0.00 | 30.11±0.01 (24h) | 1.25±0.00 | 32.20±0.12 (48h) | 0.67±0.00 |
| 22 | CTB102 | 58.20±0.11 (60h) | 0.97±0.00 | 44.01±0.23 (48h) | 0.92±0.00 | 36.12±0.19 (48h) | 0.75±0.00 |
| 23 | S6 | 64.20±0.28 (48h) | 1.34±0.01 | 53.56±0.14 (48h) | 1.12±0.00 | 37.43±0.27 (48h) | 0.78±0.01 |
| 24 | S10 | 60.00±0.85 (48h) | 1.25±0.02 | 37.56±0.06 (48h) | 0.78±0.00 | 36.27±0.04 (48h) | 0.76±0.00 |
| 25 | S11 | 64.97±0.25 (48h) | 1.35±0.01 | 57.99±0.34 (48h) | 1.21±0.01 | 37.09±0.13 (48h) | 0.77±0.00 |
| 26 | CHOB1102 | 46.52±0.28 (48h) | 0.97±0.01 | 38.59±0.23 (36h) | 1.07±0.01 | 17.63±0.21 (36h) | 0.49±0.01 |
| 27 | CHOB1103 | 60.78±0.23 (48h) | 1.27±0.00 | 57.03±0.22 (48h) | 1.19±0.00 | 29.24±0.10 (48h) | 0.61±0.00 |
| 28 | RY2305 | 51.03±0.03 (48h) | 1.06±0.00 | 35.73±0.26 (36h) | 0.99±0.01 | 19.66±0.23 (48h) | 0.41±0.00 |
| 29 | CHOB08 | 34.00±0.11 (36h) | 0.94±0.00 | 27.75±0.51 (36h) | 0.77±0.01 | 2.40±0.12 (48h) | 0.05±0.00 |
| 30 | CHOB09 | 42.30±0.57 (36h) | 1.17±0.02 | 36.14±0.09 (48h) | 0.75±0.00 | 4.60±0.18 (36h) | 0.13±0.00 |
| 31 | CTB10 | 43.04±0.20 (60h) | 0.72±0.00 | 30.24±0.59 (24h) | 1.26±0.02 | 2.67±0.05 (36h) | 0.07±0.00 |
| 32 | CTB11 | 44.10±0.17 (36h) | 1.23±0.00 | 31.99±0.15 (36h) | 0.89±0.00 | 3.84±0.21 (24h) | 0.16±0.01 |
| 33 | KK012 | 30.96±0.69 (36h) | 0.86±0.02 | 30.01±0.25 (36h) | 0.83±0.01 | 2.94±0.07 (36h) | 0.08±0.00 |
| 34 | CTB013 | 43.44±0.19 (48h) | 0.90±0.00 | 33.66±0.03 (24h) | 1.40±0.00 | 2.69±0.15 (12h) | 0.22±0.01 |
| 35 | CTB014 | 39.75±0.03 (36h) | 1.10±0.00 | 30.57±0.18 (24h) | 1.27±0.01 | 14.61±0.27 (36h) | 0.41±0.01 |
| 36 | KK015 | 37.88±0.11 (48h) | 0.79±0.00 | 33.02±0.24 (36h) | 0.92±0.01 | 18.33±0.07 (36h) | 0.51±0.00 |
| 37 | CTB016 | 39.46±0.14 (36h) | 1.10±0.00 | 30.48±0.18 (24h) | 1.27±0.01 | 14.77±0.31 (36h) | 0.41±0.01 |
| 38 | RZ017 | 39.26±0.25 (60h) | 0.65±0.00 | 29.20±0.15 (24h) | 1.22±0.01 | 14.53±0.13 (36h) | 0.40±0.00 |
| 39 | CTB018 | 33.77±0.20 (48h) | 0.70±0.00 | 31.79±0.04 (24h) | 1.32±0.00 | 17.32±0.11 (36h) | 0.48±0.00 |
| 40 | CB019 | 40.67±0.06 (60h) | 0.68±0.00 | 31.91±0.13 (36h) | 0.89±0.00 | 15.47±0.20 (36h) | 0.43±0.01 |
P, ethanol concentration; Qp, volumetric ethanol productivity.
Table 3 shows the kinetic parameters for the ethanol production at high temperatures by 6 isolates of P. kudriavzevii using SCB hydrolysate with 85g/L glucose as a substrate. The ethanol concentrations, volumetric ethanol productivities and theoretical ethanol yields obtained in this study were significantly different depending on the strain being tested. At 37 and 40°C, all isolates of the selected thermotolerant yeasts produced significantly higher ethanol concentrations, volumetric ethanol productivities and theoretical ethanol yields than those at 45°C. However, among the tested isolates, P. kudriavzevii RZ8-1 yielded relatively higher ethanol concentrations, volumetric ethanol productivities and theoretical ethanol yields than the other isolates at all tested temperatures, suggesting that this strain is a good candidate for ethanol production from SCB hydrolysate in high temperature conditions. Thus, it was selected for further study.
Kinetic parameters of ethanol production at high temperatures by the selected thermotolerant yeasts using SCB hydrolysate as a substrate.
| Strain | Temp. (°C) | Kinetic parameters (mean±SD) | ||||
|---|---|---|---|---|---|---|
| P (g/L) | Yp/s (g/g) | Qp (g/Lh) | T.Y (%) | t (h) | ||
| RZ3-1 | 37 | 32.54±0.06d | 0.38±0.00d | 1.36±0.00d | 74.92±0.13d | 24 |
| 40 | 30.78±0.02d | 0.36±0.00c | 1.28±0.00e | 70.85±0.05d | 24 | |
| 45 | 2.32±0.14a | 0.03±0.00a | 0.19±0.01a | 5.34±0.33a | 12 | |
| RZ5-1 | 37 | 33.91±0.36b | 0.40±0.00b | 1.41±0.02b | 78.06±0.83b | 24 |
| 40 | 33.43±0.15b | 0.39±0.00a,b | 1.39±0.01b | 76.95±0.34b | 24 | |
| 45 | 2.35±0.05a | 0.03±0.00a | 0.20±0.00a | 5.40±0.11a | 12 | |
| RZ8-1 | 37 | 35.51±0.03a | 0.42±0.00a | 1.48±0.01a | 81.75±0.07a | 24 |
| 40 | 33.84±0.13a | 0.40±0.00a | 1.41±0.00a | 77.91±0.29a | 24 | |
| 45 | 2.44±0.04a | 0.03±0.00a | 0.20±0.00a | 5.62±0.10a | 12 | |
| S6-1 | 37 | 33.49±0.03b,c | 0.39±0.00c | 1.40±0.01b,c | 77.10±0.07b,c | 24 |
| 40 | 32.81±0.17c | 0.39±0.00b | 1.37±0.00d | 75.54±0.39c | 24 | |
| 45 | 2.45±0.04a | 0.03±0.00a | 0.20±0.00a | 5.63±0.08a | 12 | |
| S10 | 37 | 33.07±0.08c | 0.39±0.00c | 1.38±0.00c | 76.13±0.18c | 24 |
| 40 | 33.21±0.03b | 0.39±0.00a,b | 1.38±0.00c | 76.46±0.07b | 24 | |
| 45 | 2.36±0.04a | 0.03±0.00a | 0.20±0.00a | 5.43±0.10a | 12 | |
| S11 | 37 | 35.33±0.30a | 0.42±0.00a | 1.47±0.01a | 81.34±0.68a | 24 |
| 40 | 32.74±0.21c | 0.39±0.00b | 1.36±0.01d | 75.37±0.47c | 24 | |
| 45 | 2.38±0.04a | 0.03±0.00a | 0.20±0.00a | 5.48±0.10a | 12 | |
P, ethanol concentration; Yp/s, ethanol yield; Qp, volumetric ethanol productivity; T.Y, theoretical ethanol yield; t, fermentation time; mean values±standard deviation (SD) with different letters in the same column are significant different at p<0.05 based on Duncan's Multiple Range Test (DMRT) analysis.
The effect of heat, ethanol and acetic acid stress on the growth of P. kudriavzevii RZ8-1 was determined, and the results are illustrated in Fig. 2. As shown in Fig. 2A, the growth of P. kudriavzevii RZ8-1 was unchanged when it was cultured on YM agar at 30, 37 and 40°C. A slight decrease in the growth of P. kudriavzevii RZ8-1 was observed at 42°C. P. kudriavzevii RZ8-1 can grow at temperatures up to 45°C, although its growth dramatically decreased compared to the other tested temperatures. Fig. 2B shows the effect of ethanol stress on the growth of P. kudriavzevii RZ8-1. As found in this study, P. kudriavzevii RZ8-1 grew well in the medium containing 5% ethanol when compared to the control medium without ethanol supplementation. A slight decrease in the growth of yeast cells was observed when they were cultured in media containing 8% or 10% ethanol. P. kudriavzevii RZ8-1 could grow in a medium containing 12% ethanol, but its growth was remarkably decreased compared with other conditions. Acetic acid is the most common weak acid present in SCB hydrolysate, and it is formed by the de-acetylation of hemicellulose.48,49 The effect of acetic acid stress on the cell growth of P. kudriavzevii RZ8-1 was determined, and the results are summarized in Fig. 2C. There were no significant differences in the growth of P. kudriavzevii RZ8-1 on media supplemented with 0.5, 1.0 or 2.5g/L acetic acid. In the medium supplemented with 5.0g/L acetic acid, a slight decrease in growth was observed. When the acetic acid concentration in the medium was increased to 7.5g/L, the growth of P. kudriavzevii RZ8-1 was hardly detected, similar to what was reported by Favaro et al.50
Viability test of P. kudriavzevii RZ8-1 during fermentation at high temperatureThe viability of P. kudriavzevii RZ8-1 during fermentation at high temperature (40°C) using SCB hydrolysate as a sole carbon source was determined. As found in the current study, the number of viable cells was almost constant, ranging from 2.44×108 to 3.03×108cells/mL, for at least eight successive cycles (192h). The number of dead cells was also constant throughout the entire operation, ranging from 1.35×107 to 1.65×107cells/mL. These findings suggest that this thermotolerant yeast strain can be potentially used for industrial ethanol production at high temperatures (Additional Information Section, Table 1).
Morphological characterization of the thermotolerant yeast P. kudriavzevii RZ8-1Morphological analysis of the thermotolerant yeast P. kudriavzevii RZ8-1 was carried out using bright field microscopy and scanning electron microscopy (SEM), and the results are shown in Fig. 3. Colonies of P. kudriavzevii RZ8-1 growing on YM agar medium at 35°C were tannish-white in color, dull to occasionally almost powdery, butyrous, low convex with flattened centers, and had margins varying from smooth to lobed. Cells of P. kudriavzevii RZ8-1 became ovoid to elongated and occurred singly or in pairs when grown in YM broth at 35°C. SEM analysis revealed that this strain had a cellular size of approximately 60μm in length and 25μm in diameter. In addition, it also showed multilateral budding similar to what has been observed in other yeast species, such as S. cerevisiae and K. marxianus.
Ethanol fermentation and gene expression analysis using qRT-PCRThe results of ethanol production by the selected thermotolerant yeast P. kudriavzevii RZ8-1 using a YM medium containing 100g/L glucose at 30 and 42°C are summarized in Additional Information Section, Table 2. As found in the current study, P. kudriavzevii RZ8-1 produced greater ethanol concentration and volumetric ethanol productivity at 30°C than that at 42°C. In addition, the number of viable cells at 30°C was also greater than that at 42°C. The highest ethanol concentrations and volumetric ethanol productivities produced by this strain were 44.63g/L and 1.57g/Lh at 30°C, and 38.01g/L and 1.39g/Lh at 42°C.
The expression levels of genes encoding HSPs (hsp90 and ssq1), neutral trehalase (nth1 606 and nth1572), trehalose synthase (ggs1), alcohol dehydrogenase (adh1, adh2, adh3 and adh4), glycogen synthase kinase-3 (gsk3), glyceraldehyde-3-phosphate dehydrogenase (tdh2) and enolase (eno) in the thermotolerant yeast P. kudriavzevii RZ8-1 during ethanol fermentation at 30 and 42°C were examined using qRT-PCR. The results are summarized in Table 4. As found in this study, the expression of hsp90 and ssq1 was induced under long-term heat stress conditions. However, their expression levels decreased under heat shock treatment compared to control and long-term heat stress conditions. With respect to the genes involved in trehalose metabolism (nth1 606, nth1572 and ggs1), the transcription levels of all genes decreased under both long-term heat stress and heat shock conditions compared with control condition.
Gene expression analysis in P. kudriavzevii RZ8-1 during ethanol fermentation at high temperature (42°C) by quantitative real-time polymerase chain reaction (qRT-PCR).
| Genes | qRT-PCR change (n-fold)a | ||
|---|---|---|---|
| 30°C | Long-term heat stress | Heat shock condition | |
| hsp90 | 0.59±0.07d | 2.69±0.41a | 0.17±0.01c |
| ssq1 | 0.81±0.16d | 2.76±0.27a | 0.58±0.17b |
| nth1 606 | 4.19±0.27d | 0.82±0.26b | 1.00±0.04 |
| nth1572 | 3.53±0.14d | 0.34±0.10b,c,d | 1.00±0.16a |
| ggs1 | 11.87±1.01c | 0.91±0.14b | 1.00±0.19a |
| adh1 | 0.05±0.01d | 0.65±0.12b,c | 1.10±0.15a |
| adh2 | 0.35±0.05d | 0.19±0.02c,d | 1.01±0.17a |
| adh3 | 0.25±0.05d | 0.70±0.05b,c | 1.05±0.20a |
| adh4 | 0.07±0.03d | 0.05±0.01d | 1.15±0.07a |
| gsk3 | 39.82±1.16b | 0.83±0.04b | 1.00±0.23a |
| tdh2 | 1.59±0.01d | 2.66±0.28a | 1.00±0.08a |
| eno | 124.80±9.40a | 2.58±0.69a | 0.45±0.09b,c |
The expression of all genes encoding alcohol dehydrogenases in P. kudriavzevii RZ8-1 (adh1, adh2, adh3 and adh4) was up-regulated under heat shock conditions. However, the transcription levels of only two genes, i.e., adh1 and adh3, greatly increased under long-term heat stress compared to the control condition, whereas the transcription levels of the other two genes, i.e., adh2 and adh4, slightly decreased under such conditions in comparison to the control. The expression levels of eno and gsk3 in P. kudriavzevii RZ8-1 were reduced under both long-term heat stress and heat shock conditions when compared with the control treatment; a similar result was found for the genes involved in trehalose metabolism. These results suggested that both short- and long-term heat stress suppressed the expression of eno and gsk3 genes in P. kudriavzevii RZ8-1. With respect to tdh2, expression was induced under long-term heat stress, and it was slightly decreased under heat shock conditions.
DiscussionHTEF provides several advantages over low temperature ethanol fermentation (LTEF).1,2 One of the keys for successful HTEF is the utilization of a high potential thermotolerant yeast that is capable of growth and ethanol production at high temperatures. In this study, thermotolerant ethanol-producing yeasts were successfully isolated from plant orchard samples and were cultivated at high temperatures using an enrichment medium supplemented with 4% (v/v) ethanol. This enrichment culture technique has been widely used by several researchers to isolate thermotolerant yeasts. For example, Limtong et al.3 reported the isolation of thermotolerant yeasts at 35°C using a YM medium supplemented with 4% (v/v) ethanol and found several thermotolerant yeast strains that were capable of growth and ethanol production at high temperatures. Among the isolated thermotolerant yeasts, K. marxianus DMKU 3-1042 was the most effective strain for ethanol production at a high temperature of 45°C. Kumar et al.51 reported the isolation of thermotolerant yeasts from soil samples collected from the dumping sites of crushed sugarcane bagasse at a sugar mill using enrichment culture at 50°C. They found one strain of yeast, designated Kluyveromyces sp. IIPE453, that exhibits a high efficiency of growth and ethanol fermentation at the high temperatures of 45 and 50°C. Charoensopharat et al.5 reported the isolation of thermotolerant yeasts from soil and plant materials collected from Jerusalem artichoke (JA) plantations using a YM medium supplemented with 4% (v/v) ethanol and incubation at 35°C. They found six isolates of thermotolerant yeasts that were capable of growth and ethanol production at high temperatures of up to 45°C. Among the isolated thermotolerant yeast strains, K. marxianus DBKKU Y-102 was found to be the most effective strain for direct ethanol fermentation at a high temperature (40°C) using JA tubers as raw material under consolidated bioprocessing. Recently, Nuanpeng et al.2 reported the isolation of thermotolerant yeasts from soil and plant materials collected from sugarcane and sweet sorghum plantations using a YM medium supplemented with 4% (v/v) ethanol and incubation at 35°C. The authors obtained several potential thermotolerant yeasts that were capable of growth and ethanol production at 40 and 45°C. Among the isolated strains, a newly isolated thermotolerant yeast, designated S. cerevisiae DBKKU Y-53, exhibited great potential for ethanol production from sweet sorghum juice at 40°C.
Using an enrichment culture technique, 127 isolates of yeasts were obtained in the present study. Among the isolated yeast strains, only 40 isolates were classified as thermotolerant fermentative yeast since they could ferment glucose and accumulate relatively high levels of CO2 in the fermentation medium. The accumulation of CO2 in a fermentation medium containing glucose as a carbon and energy source has been successfully used for the preliminary screening of ethanol fermentative yeasts.2,5 To identify the selected thermotolerant yeasts obtained in this study, sequencing analysis of the D1/D2 domain of the large subunit of the 26S rDNA gene was performed. This technique has been widely used to identify several yeast strains, e.g., S. cerevisiae,2K. marxianus5 and P. kudriavzevii.12 Based on sequencing analysis of the D1/D2 region, six yeast species were identified in the present study, i.e., P. kudriavzevii, C. tropicalis, C. grabrata, C. albicans, C. orthopsilosis and K. ohmeri (Fig. 1). Several strains of the isolated thermotolerant yeasts identified in this work, particularly C. tropicalis, C. grabrata, C. albicans, and C. orthopsilosis, have been reported as medical pathogens, but they are also good candidates for ethanol production from different feedstocks, such as Avicel PH101 microcrystalline cellulose,52 agro-residues,53 non-detoxified acid pretreated corncob,54 starch55 and cellulose.56 Although K. ohmeri has been reported as a rare yeast pathogen and has recently emerged as an important cause of fungemia in immunocompromised patients, it exhibits a great potential for ethanol production from glucose.57P. kudriavzevii has been reported as an effective yeast strain for high-temperature ethanol production from sugarcane juice,7 cornstalk hydrolysate9 and cassava hydrolysate.12
The ethanol production efficiency of 40 isolates of putative thermotolerant yeasts was evaluated at high temperatures using glucose as a carbon source, and the results found that the maximum ethanol concentrations and volumetric ethanol productivities were significantly different depending on the yeast strain and fermentation temperatures. Out of the 40 isolates of the selected thermotolerant yeasts, 6 isolates of P. kudriavzevii, i.e., RZ3-1, RZ5-1, RZ8-1, S6, S10 and S11, exhibited great potential for ethanol production at 37, 40 and 45°C compared to the other tested isolates (Table 2). The highest ethanol concentrations and volumetric ethanol productivities were produced by the newly isolated thermotolerant yeast P. kudriavzevii; these results were greater than those from K. marxianus IBM3 using glucose at 160g/L as a substrate under the same fermentation conditions at 37, 40 and 45°C.58 In addition, the ethanol production efficiency of P. kudriavzevii was also comparable with that of S. cerevisiae VS3.47 Among the 6 isolates of P. kudriavzevii, isolate RZ8-1 was found to be the most promising strain for ethanol production at high temperatures using SCB hydrolysate as a substrate. The maximum ethanol concentrations and volumetric ethanol productivities of P. kudriavzevii RZ8-1 at high temperatures (37 and 40°C) were greater than those of P. kudriavzevii HOP-1 at 30°C using cotton stalks hydrolysate as substrate,42 and S. cerevisiae at 35°C using rapeseed straw hydrolysate as feedstock.59 Although the highest ethanol concentrations produced by P. kudriavzevii RZ8-1 at high temperatures were lower than that of I. orientalis IPE100 at 42°C, the volumetric ethanol productivities of this thermotolerant yeast (1.48 and 1.41g/Lh at 37 and 40°C, respectively) were greater than that of I. orientalis IPE100 (0.96g/Lh).9 It should be noted from the current study that the highest ethanol concentrations and volumetric ethanol productivities of P. kudriavzevii RZ8-1 at 40 and 45°C were lower than those of P. kudriavzevii DMKU 3-ET15, but this is due to their differences in the raw materials and initial sugar concentrations used in the fermentation process. In the case of P. kudriavzevii DMKU 3-ET15, the authors used cassava starch hydrolysate containing 180g/L glucose as a substrate,12 whereas SCB hydrolysate containing 85g/L glucose was used in this study for ethanol production by P. kudriavzevii RZ8-1.
During ethanol fermentation, yeast cells may encounter not only heat and ethanol stresses but also other stresses (e.g., acetic acid) that have a negative effect on cell growth and ethanol production, particularly when the hydrolysate of lignocellulosic materials is used as a substrate. Acetic acid has been reported to cause a decrease in the cytoplasmic pH and inhibit the activity of several enzymes, such as endolase, phosphoglyceromutase, aldolase and triosephosphate isomerase, resulting in the reduction of cell growth and ethanol production efficiency.60–65 The present study tested the effects of heat-, ethanol- and acetic acid stress on cell growth in the newly isolated thermotolerant yeast P. kudriavzevii RZ8-1. As found in this work, P. kudriavzevii RZ8-1 exhibited multi-stress tolerance toward heat, ethanol and acetic acid stress (Fig. 2), which is similar to P. kudriavzevii MF-121.13,14 These findings suggested that P. kudriavzevii RZ8-1 might have its own adaptive response to stressful conditions by either changing its cell membrane composition or synthesizing a group of proteins known as HSPs. Ohta et al.66 reported that high temperature conditions cause an increase in cell membrane fluidity, and yeasts respond to this physical change by changing their fatty acid composition. In addition, increases in growth temperature usually lead to the synthesis of HSPs, which play an important role in conferring thermal and ethanol cross-tolerance in various microorganisms, such as S. cerevisiae and K. marxianus.2,5,67,68 Wei et al.69 reported that high membrane integrity and intracellular catalase activity were involved in the ability of C. krusei S4-3 to tolerate acetic acid. It should be noted that the ability of yeast cells to tolerate high temperatures, high ethanol concentrations or high acetic acid concentrations depends on several factors, including the composition of the fermentation medium, intracellular ethanol accumulation, incubation temperature, osmotic pressure and yeast strain.70,71
Previous studies in S. cerevisiae and K. marxianus revealed that heat stress during ethanol fermentation stimulates the expression of several stress-responsive genes encoding HSPs and the proteins involved in the ethanol production pathway and other metabolic processes.2,5,31,67 To the best of our knowledge, there is less information regarding the heat stress response of thermotolerant P. kudriavzevii during ethanol production at high temperatures. In this study, the expression levels of genes encoding HSPs, neutral trehalase, trehalose synthase, alcohol dehydrogenase, glycogen synthase kinase-3, glyceraldehyde-3-phosphate dehydrogenase and enolase were evaluated (Table 4). Although the expression levels of hsp90 and ssq1, which encode HSP90 and HSP70 family ATPase, respectively, were low under short-term stress or heat shock stress at 42°C compared to the control condition without stress treatment (30°C), their expression levels were up-regulated under long-term heat stress at 42°C. Our results were similar to Auesukaree et al.,67 who reported high expression levels of the genes encoding the small HSPs, HSP70, HSP90 and HSP100 family in S. cerevisiae under heat shock and long-term heat exposure at 37°C. Nuanpeng et al.2 also reported high expression levels of the genes encoding HSP26, HSP70, HSP90 and HSP104 in S. cerevisiae DBKKU Y-53 during ethanol fermentation at 40°C. Several HSPs, such as HSP60, HSP70, HSP90 and HSP104, are constitutively expressed at appropriate temperatures and play a crucial role as molecular chaperones by helping with protein folding and assembly, refolding of damaged proteins, stabilization of new proteins and protein transport across the membranes within the cells. In addition, some of the HSPs also possess a proteolytic activity to prevent the accumulation of the denatured proteins inside the cells.72 The high expression levels of hsp90 and ssq1 under long-term heat stress observed in this study suggested that these genes may play a crucial role in P. kudriavzevii RZ8-1 during ethanol fermentation at 42°C. To clarify the precise biological functions of hsp90 and ssq1 in P. kudriavzevii RZ8-1, further study, such as gene disruption, is needed.
It should be noted that the expression of genes encoding enolase and the enzymes involved in trehalose and glycogen metabolism were up-regulated under non-stress conditions and were down-regulated under both heat shock and long-term heat exposure at 42°C in this study. Enolase is a phosphopyruvate hydratase that catalyzes the conversion of 2-phosphoglycerate to phosphoenolpyruvate during glycolysis and catalyzes the reverse reaction during gluconeogenesis.73 In S. cerevisiae, eno was up-regulated during co-fermentation of glucose and xylose74 as well as in response to galactose induction,75 which suggested that it was involved in sugar metabolism. In addition, eno was also highly expressed in S. cerevisiae during exposure to a high temperature at 40°C,2 which is different from this study, in which its expression level in P. kudriavzevii RZ8-1 decreased upon exposure to heat shock and long-term heat stress at 42°C. There is no reason for these different expression levels between the two yeast strains, but based on a study of Edwards et al.,76eno may have alternative functions in the cell wall that could be a heat-related effect. The cell wall of S. cerevisiae reacts differently than P. kudriavzevii by changing its expression of eno.73 Thus, further study is necessary to clarify this phenomenon in thermotolerant P. kudriavzevii RZ8-1. In the case of trehalose and glycogen, it has been reported that these two reserve carbohydrates are implicated in the survival of yeast cells under various stress conditions, such as heat, ethanol, osmotic and oxidative stresses. Trehalose and glycogen have been reported to play key roles as membrane protectants, compatible solutes and reserve carbohydrates that can be mobilized during stress.77,78 They are also involved in energy homeostasis, which is essential to withstanding adverse environmental conditions and maintaining cellular integrity.79 Bandara et al.78 reported that trehalose promotes the survival of S. cerevisiae during lethal ethanol stress, but it does not play a role in ethanol tolerance at sublethal ethanol concentrations. In S. cerevisiae, the expression levels of genes encoding trehalose synthase and glycogen synthase increased upon exposure to a mild heat stress and salt shock, while those encoding the neutral trehalase decreased upon heat stress conditions.34 With respect to the current study, the expression levels of all genes encoding trehalose synthase (ggs1), neutral trehalase (nth1606 and nth1572) and glycogen synthase kinase-3 (gsk3) in P. kudriavzevii RZ8-1 decreased upon exposure to heat shock and long-term heat stress at 42°C compared with the control conditions. These findings suggested that a high temperature at 42°C suppressed the expression of genes involved in trehalose and glycogen metabolism in P. kudriavzevii RZ8-1. It should be noted from the current study that the high expression level of the gsk3 gene under non-stress conditions may be related to the growth of yeast cells. Previous studies demonstrated that glycogen synthase kinase-3 family members play key roles in nitrogen-responsive phosphorylation of the meiotic regulator Ume6p,80 cell cycle regulation81 and stress responses.82 Furthermore, it has also been reported that the gsk3 gene is involved in the regulation of ribosome biogenesis in yeast cells.83
The gene encoding alcohol dehydrogenase has been cloned and characterized from many different yeast strains, such as P. stipites,84S. cerevisiae85 and K. marxianus.86,87 There are two (i.e., Psadh1 and Psadh2), seven (i.e., adh1 to adh7) and four genes (i.e., adh1 to adh4) encoding alcohol dehydrogenases in P. stipitis, S. cerevisiae and K. marxianus, respectively. The transcriptional activation of these genes varies depending on the yeast strains, growth phases and environmental conditions. In K. marxianus, for example, adh1 and adh2 are highly expressed in the exponential phase at 30°C compared with adh3 and adh4. In contrast, adh3 and adh4 are largely expressed in the stationary phase at 30°C, and their expression levels are largely induced by heat stress at 45°C.5,86 There is less information on the expression of adh genes in P. kudriavzevii during ethanol fermentation at high temperatures. In the present study, the transcriptional levels of four of the adh genes in P. kudriavzevii RZ8-1 were investigated, and the results revealed that their expression levels increased upon exposure to heat stress at 42°C, which is similar to the results reported by Lertwattanasakul et al.86 and Charoensopharat et al.5 These findings suggested that the adh genes in P. kudriavzevii RZ8-1 were up-regulated at a high temperature.
With respect to the tdh2 gene encoding glyceraldehyde-3-phosphate dehydrogenase in P. kudriavzevii, its expression level greatly increased upon exposure to long-term heat stress at 42°C, similar to what was reported by Charoensopharat et al.5 In S. cerevisiae and C. albicans, the genes encoding glyceraldehyde-3-phosphate dehydrogenase are largely expressed in the stationary growth phase and are involved in glycolysis and gluconeogenesis.88 In K. marxianus, the tdh2 gene is highly expressed in both the exponential and stationary phases upon exposure to heat stress at 45°C.5 Based on the high expression level of the tdh2 gene in P. kudriavzevii RZ8-1 upon heat stress at 42°C, we propose that this gene encodes a heat shock protein, and it may also be involved in glycolysis and gluconeogenesis in this organism.
ConclusionsBased on the results of the current study, a newly isolated yeast, P. kudriavzevii RZ8-1, was the most effective thermotolerant yeast strain for ethanol production at high temperatures. It exhibited good growth and ethanol production capability at temperatures up to 45°C. The maximum ethanol concentrations produced by P. kudriavzevii RZ8-1 were 59.47, 69.85 and 35.09g/L at 37, 40 and 45°C, respectively, using glucose at 160g/L as a carbon source. When SCB hydrolysate containing 85g/L glucose was used as a substrate, this strain produced the highest ethanol concentrations of 35.51, 33.84 and 2.44g/L at 37, 40 and 45°C, respectively. It also exhibited multi-stress tolerance toward heat, ethanol and acetic acid stresses. Furthermore, it viability was also high throughout the entire operations of repeated batch fermentation at 40°C. During ethanol fermentation at 42°C, genes encoding HSP70 (ssq1), HSP90 (hsp90), alcohol dehydrogenase (adh1, adh2, adh3 and adh4) and glyceraldehyde-3-phosphate dehydrogenase (tdh2) were up-regulated, whereas those encoding enolase (eno), trehalose synthase (ggs1), neutral trehalase (nth1606 and nth1572) and glycogen synthase kinase-3 (gsk3) were down-regulated upon exposure to heat stress at 42°C. These findings suggested that the high-temperature growth and ethanol fermentation activities of the newly isolated thermotolerant yeast P. kudriavzevii RZ8-1 were closely correlated to the expression of genes involved in heat shock response and the ethanol production pathway.
Conflicts of interestThe authors declare no conflicts of interest.
This work was financially supported by the Research Center for Alternative Energy Research and Development (AERD), Khon Kaen University, and the Energy Policy and Planning Office, Ministry of Energy, Royal Thai Government.