To test the hypothesis that the pattern of gene expression in circulating leukocytes may differ between vascular compartments, depending on the presence or absence of atherosclerosis, we evaluated the regional vascular differences in patterns of inflammatory cell activation.
MethodsPatients (n=8) with angiographically-established coronary artery disease (CAD+) and 8 without (CAD−) had blood samples taken from a peripheral vein as well as from left and right coronary arteries. Samples were pooled resulting in 4 CAD+ samples versus 4 CAD− samples and hybridised to a Whole Human Genome Microarray 4×44K.
ResultsCAD− patients had a similar gene expression profile across the different sites. CAD+ patients had statistically significant different gene expression patterns in venous vs. right and left coronary artery compartments. The expression pattern observed in the right coronary was where the most differences in gene expression were observed in CAD+ vs. CAD− patients. Overall, 1964 genes were differentially expressed between CAD+ and CAD−. Of these, 1052 were less expressed in CAD+ and 912 were more expressed in CAD+. Up to 12 of the 20 most differentially expressed genes appeared to reflect different phases of the atherosclerosis process: endothelial dysfunction, lipid accumulation, and smooth muscle cell proliferation.
ConclusionsGene expression of circulating leukocytes differentiates CAD+ from CAD− patients. Gene expression is significantly different between coronary arteries and the systemic circulation in CAD+ patients, but not in CAD− patients. Gene expression is significantly different between CAD+ and CAD− subjects, and appears to reflect the atherosclerosis process. These intra-individual differences may be an additional feature of established coronary artery disease.
Para comprobar la hipótesis de que los patrones de expresión génica de leucocitos en circulación pueden ser diferentes entre los compartimentos vasculares dependiendo de la presencia o ausencia de arteriosclerosis, hemos evaluado en distintas regiones vasculares las diferencias entre los patrones de expresión y la activación de células inflamatorias.
MétodosSe extrajeron muestras de sangre de venas periféricas y de las arterias coronarias (derecha e izquierda) de pacientes con (n=8; CAD+) y sin (n=8; CAD−) enfermedad arterial coronaria establecida angiográficamente. Las muestras fueron hibridadas en dos pooles de 4 muestras (CAD+ vs CAD−) mediante el kit Whole Human Genome Microarray 4×44K.
ResultadosLos pacientes CAD− tenían un perfil de expresión génica similar entre los distintos compartimentos vasculares. Los pacientes CAD+ tenían patrones de expresión génica significativamente diferentes entre los compartimentos venosos y las arterias coronarias derecha e izquierda. El patrón de expresión observado en la arteria coronaria derecha fue el que presentó más diferencias entre los pacientes CAD+ vs. CAD−. En conjunto, 1.964 genes estaban expresados diferencialmente entre CAD+ y CAD−. De estos, 1.052 estaban menos expresados en CAD+ i 912 estaban más expresados en CAD+. Hasta 12 de los 20 genes más diferencialmente expresados estaban relacionados con las diferentes fases del proceso arteriosclerótico: disfunción endotelial, acumulación lipídica y proliferación de células musculares lisas.
ConclusionesLa expresión génica de leucocitos circulantes diferencia pacientes CAD+ de CAD−. La expresión genética es significativamente diferente entre arterias coronarias y circulación sistémica en pacientes CAD+, pero no en pacientes CAD−. Estas diferencias intraindividuales podrían ser una característica adicional en el diagnóstico de la enfermedad arterial coronaria.
Atherosclerosis is an inflammatory disease.1 The mechanisms involved include activation of the endothelium, leukocytes and complement, as well as the generation of oxidative stress. Such activation can be detected through RNA expression analyses of circulating cells, and has been widely investigated as a marker of atherosclerosis.2–5
Some studies have described the degree of inflammation as being higher in the coronary arteries than in peripheral veins in patients with unstable angina.6,7 In line with this, we have recently described an inflammatory gradient of myeloperoxidase (MPO) decreasing from the peripheral veins towards the coronaries in patients with stable angina,8 confirming that there are regional differences with respect to leucocyte activation.
Since regional differences may also be detected at the level of RNA expression. We explored the possibility that the expression of genes in circulating cells in the coronary arteries may differ from that of peripheral blood cells.
To address this hypothesis we analysed global RNA expression from leukocytes obtained from peripheral veins and compared it to that from leukocytes from the coronary artery blood to test whether regional differences in expression would enable the differentiation of patients with coronary artery disease (CAD) from those without.
MethodsParticipantsParticipant selection and sample collection have been published elsewhere.8 Briefly, subjects who visited the outpatient clinic of the Department of Cardiology of the Sint Franciscus Gasthuis (Rotterdam, The Netherlands) and who were scheduled to undergo a diagnostic coronary angiography were asked to participate. Subjects were divided into 2 groups. The first consisted of patients in whom CAD was clinically suspected but was ruled-out based on the subsequent coronary angiography. These subjects, who constituted the control group, did not have a previous history of peripheral or cerebral vascular disease. The second group consisted of subjects with, at least, wall irregularities in left coronary (LC) and right coronary (RC) systems. Wall irregularities were defined as plaques observed in the coronaries and occupying <50% diameter (stenosis) of the lumen. The coronary angiographs were evaluated by an independent cardiologist who was blinded with respect to the provenance of the subjects.
Exclusion criteria were the presence of inflammatory disorders including rheumatoid arthritis, systemic lupus erythematous and infections, plasma C-reactive protein (CRP) >10mg/L, and disorders of kidney, liver and thyroid.
The Independent Ethics Committee of the St. Franciscus Gasthuis in Rotterdam and the Regional Independent Medical Ethical Committee at the Maasstad Hospital in Rotterdam approved the study. The participants gave written informed consent to participation in the study.
Just before angiography, a venous blood sample was taken from a peripheral vein of the forearm. Blood samples were obtained from each coronary artery in the course of the angiography.
For the extraction of RNA needed for the microarray experiments, 3mL whole blood was drawn from each site into Tempus® blood RNA tubes (Applied Biosystems). RNA samples from 8 patients with CAD and 8 without CAD were pooled 2 by 2, resulting in 4 groups of CAD− and CAD+ individuals.
RNA isolationThe extraction of total RNA was performed on the ABI PRISM 6100 automated nucleic acid prep-station (Applied Biosystems) with the Tempus® 12-port RNA isolation kit according to the manufacturer's instructions. Before micro-array analysis was performed, RNA integrity was checked with the Experion automated electrophoresis system (Bio-Rad) using the RNA highSens analysis chip (Bio-Rad).
Micro-array data generation and analysisOne-colour Microarray-based Gene Expression Analysis Protocol (Agilent Technologies, Palo Alto, CA, USA) was used to amplify and label RNA. Samples were hybridised to a Whole Human Genome Microarray 4×44K (G4122F, Agilent Technologies). Cy3-labelled aRNA (1.65μg) were hybridised for 17h at 65°C in an Agilent hybridisation oven (G2545A, Agilent Technologies) set to 10rpm in a final concentration of 1×GE×hybridisation buffer HI-RPM (Agilent Technologies). Arrays were scanned at 5mm resolution on an Agilent DNA Microarray Scanner (G2565BA, Agilent Technologies) using the default settings for 4×44K format one-colour array. Images generated by the scanner were analysed using Feature Extraction software v10.1.1.1 (Agilent Technologies).
For the RNA analyses, we focused on 8 patients with CAD (all of whom were found to have multi-vessel disease) and 8 without CAD. Pool 4 from the CAD− group was identified as an outlier based on hierarchical clustering and PCA analysis and, so, was excluded from the subsequent analyses. Hence, all results refer to 3 pools of CAD− patients and 4 pools of CAD+ patients. For each pool of subjects we focused on comparing venous (VN) vs. left coronary (LC) vs. right coronary (RC) samples. Hence, 21 slides where hybridised and the following comparisons were performed: 1) differences across all sites for CAD+ patients vs. CAD− patients; 2) for each site, differences between CAD+ vs. CAD− patients; 3) differences between CAD+ vs. CAD−.
Statistical analysesFor the micro-array analysis, the threshold was 1, and quantiles normalised9 using the GeneSpring software. Data were log2 transformed. Default flags were considered as absent, except saturated spots that were flagged as marginal. From the initial 41081 probes present in the Agilent 4×44K chip, 26644 remained after applying three types of filter (on the 6 conditions site*CAD): (i) by expression, we retained those genes with at least 75% of the replicates in range in 3/6 conditions; (ii) by flags, we retained only those genes with at least 75% acceptable values in all conditions; and (iii) by error, we retained only those genes with CV<50% within the replicates for every condition. A mixed linear model with two fixed covariates (CAD+/− and site-of-extraction) and one random effect (pools) was fitted to the data, and estimates were obtained using methodology specifically designed for the analysis of high-throughput genomic data10 as implemented in the Limma bioconductor package (http://www.bioconductor.org/).11P-values were adjusted to control for false discovery rate (FDR) using the Benjamini-Hochberg correction.12 Functional analysis was performed using ingenuity pathway analysis. P values<0.05 (2-tailed) were considered statistically significant.
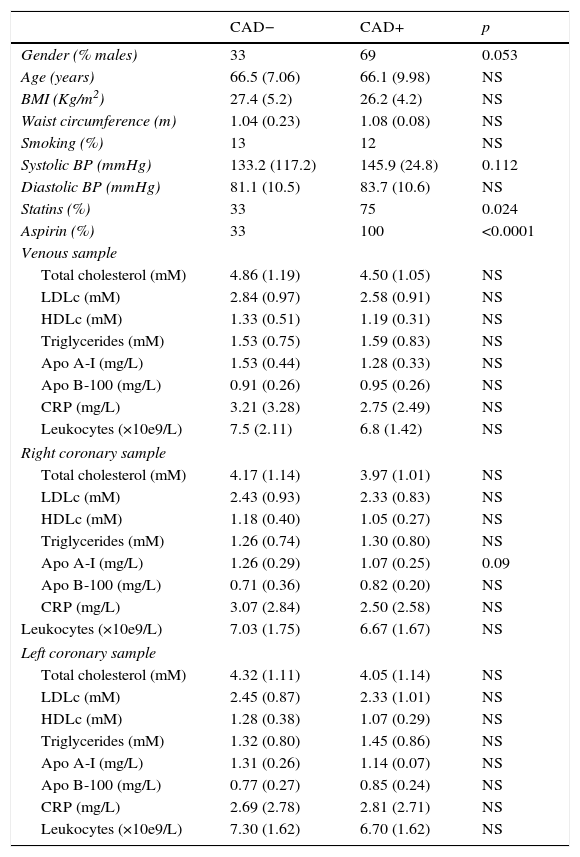
ResultsThe characteristics of the CAD− and CAD+ groups are summarised in Table 1. The CAD+ group had more males. This group also showed higher systolic blood pressure, and more subjects were receiving treatment that included statins or aspirin.
Clinical and biochemical characteristics of the study subjects.
| CAD− | CAD+ | p | |
|---|---|---|---|
| Gender (% males) | 33 | 69 | 0.053 |
| Age (years) | 66.5 (7.06) | 66.1 (9.98) | NS |
| BMI (Kg/m2) | 27.4 (5.2) | 26.2 (4.2) | NS |
| Waist circumference (m) | 1.04 (0.23) | 1.08 (0.08) | NS |
| Smoking (%) | 13 | 12 | NS |
| Systolic BP (mmHg) | 133.2 (117.2) | 145.9 (24.8) | 0.112 |
| Diastolic BP (mmHg) | 81.1 (10.5) | 83.7 (10.6) | NS |
| Statins (%) | 33 | 75 | 0.024 |
| Aspirin (%) | 33 | 100 | <0.0001 |
| Venous sample | |||
| Total cholesterol (mM) | 4.86 (1.19) | 4.50 (1.05) | NS |
| LDLc (mM) | 2.84 (0.97) | 2.58 (0.91) | NS |
| HDLc (mM) | 1.33 (0.51) | 1.19 (0.31) | NS |
| Triglycerides (mM) | 1.53 (0.75) | 1.59 (0.83) | NS |
| Apo A-I (mg/L) | 1.53 (0.44) | 1.28 (0.33) | NS |
| Apo B-100 (mg/L) | 0.91 (0.26) | 0.95 (0.26) | NS |
| CRP (mg/L) | 3.21 (3.28) | 2.75 (2.49) | NS |
| Leukocytes (×10e9/L) | 7.5 (2.11) | 6.8 (1.42) | NS |
| Right coronary sample | |||
| Total cholesterol (mM) | 4.17 (1.14) | 3.97 (1.01) | NS |
| LDLc (mM) | 2.43 (0.93) | 2.33 (0.83) | NS |
| HDLc (mM) | 1.18 (0.40) | 1.05 (0.27) | NS |
| Triglycerides (mM) | 1.26 (0.74) | 1.30 (0.80) | NS |
| Apo A-I (mg/L) | 1.26 (0.29) | 1.07 (0.25) | 0.09 |
| Apo B-100 (mg/L) | 0.71 (0.36) | 0.82 (0.20) | NS |
| CRP (mg/L) | 3.07 (2.84) | 2.50 (2.58) | NS |
| Leukocytes (×10e9/L) | 7.03 (1.75) | 6.67 (1.67) | NS |
| Left coronary sample | |||
| Total cholesterol (mM) | 4.32 (1.11) | 4.05 (1.14) | NS |
| LDLc (mM) | 2.45 (0.87) | 2.33 (1.01) | NS |
| HDLc (mM) | 1.28 (0.38) | 1.07 (0.29) | NS |
| Triglycerides (mM) | 1.32 (0.80) | 1.45 (0.86) | NS |
| Apo A-I (mg/L) | 1.31 (0.26) | 1.14 (0.07) | NS |
| Apo B-100 (mg/L) | 0.77 (0.27) | 0.85 (0.24) | NS |
| CRP (mg/L) | 2.69 (2.78) | 2.81 (2.71) | NS |
| Leukocytes (×10e9/L) | 7.30 (1.62) | 6.70 (1.62) | NS |
Lipids, CRP and leucocyte counts were comparable between CAD− and CAD+ for the three compartments studied, namely peripheral vein, right coronary artery and left coronary artery.
Genes differentially expressed across sites in CAD+ and CAD− patientsFor CAD− patients, the gene expression profile was comparable across the different sites. However, for CAD+ patients, the venous compartment displayed a significantly different behaviour in terms of gene expression, compared with the right and left coronary arteries; both of the latter sites showing a similar RNA expression pattern. The analysis of the differentially expressed genes did not segregate with any specific metabolic process, or altered pathway.
The right coronary was the location where most differences in gene expression between CAD+ and CAD− patients were found; 6 probes corresponding to 4 genes were differentially expressed between CAD+ and CAD− at all sites. These genes were RGPD1, GCC2, the class A scavenger receptor MARCO and FMO1 which is involved in the oxidative metabolism of certain drugs.
Genes differentially expressed between CAD+ and CAD−There were 1964 genes found to be differentially expressed between CAD+ and CAD− patients (α≤0.05) after multiple testing corrections to control for FDR across all genes.
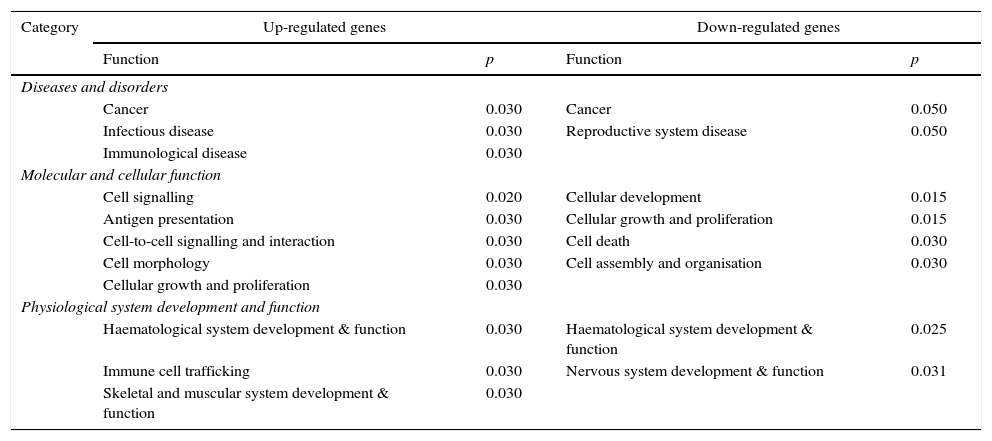
Of these genes, 1052 were more expressed in CAD− than in CAD+ patients, and 912 were more expressed in CAD+ than in CAD− patients. The biological processes that may be significantly altered as a consequence are depicted in Table 2.
Functional analyses of the main biological processes involving genes differentially expressed in CAD+ versus CAD−.
| Category | Up-regulated genes | Down-regulated genes | ||
|---|---|---|---|---|
| Function | p | Function | p | |
| Diseases and disorders | ||||
| Cancer | 0.030 | Cancer | 0.050 | |
| Infectious disease | 0.030 | Reproductive system disease | 0.050 | |
| Immunological disease | 0.030 | |||
| Molecular and cellular function | ||||
| Cell signalling | 0.020 | Cellular development | 0.015 | |
| Antigen presentation | 0.030 | Cellular growth and proliferation | 0.015 | |
| Cell-to-cell signalling and interaction | 0.030 | Cell death | 0.030 | |
| Cell morphology | 0.030 | Cell assembly and organisation | 0.030 | |
| Cellular growth and proliferation | 0.030 | |||
| Physiological system development and function | ||||
| Haematological system development & function | 0.030 | Haematological system development & function | 0.025 | |
| Immune cell trafficking | 0.030 | Nervous system development & function | 0.031 | |
| Skeletal and muscular system development & function | 0.030 | |||
The network analyses of the up-regulated genes among CAD+ patients identified potential toxicity linked to the cholesterol biosynthesis pathway. In line with this, the LDLR gene was more expressed in the CAD+ group (log2FC: 0.77; p=0.025).
Genes most up-regulated (more expressed) or down-regulated (less expressed)We identified the 20 genes (10 up-regulated, 10 down-regulated) which were most significantly differentially expressed between CAD+ and CAD−. These are listed in Table 3, and which are associated with key steps of the atherosclerotic process including endothelial dysfunction, lipoprotein accumulation and smooth muscle cell proliferation.
Top 10 genes differentially up-regulated and down-regulated in CAD+ compared with CAD− patients.
| Gene acronym | Gene name | Log2FC |
|---|---|---|
| Top 10 up-regulated | ||
| MSR1 | Macrophage scavenger receptor 1 | 1.62 |
| SCGB3A1 | Secretoglobin 3A | 1.58 |
| HLA-DQA1 | Major histocompatibility complex, class II, DQ alpha 1 | 1.57 |
| DSP | Desmoplakin | 1.44 |
| HLA-DQB1 | Major histocompatibility complex, class II, DQ beta 1 | 1.29 |
| SLC7A8 | Solute carrier family 7 (amino acid transporter, L-type), member 8 | 1.27 |
| TCOF1 | Treacher Collins–Franceschetti syndrome 1 | 1.26 |
| KIF26A | Kinesin family member 26A | 1.22 |
| RAI2 | Retinoic acid induced 2 | 1.20 |
| MRPL43 | Mitochondrial ribosomal protein L43 | 1.19 |
| Top 10 down-regulated | ||
| NKX2-3 | NK2 transcription factor related | −2.89 |
| LALBA | Lactalbumin, alpha- | −2.85 |
| CDK5R2 | Cyclin-dependent kinase 5, regulatory subunit 2 (p39) | −2.60 |
| DAB1 | Disabled homolog 1 | −2.55 |
| CRHR1 | Corticotropin releasing hormone receptor 1 | −2.53 |
| PRIMA1 | Proline rich membrane anchor 1 | −2.52 |
| MED24 | Mediator complex subunit 24 | −2.50 |
| PDLIM4 | PDZ and LIM domain 4 | −2.49 |
| WDTC1 | WD and tetratricopeptide repeats 1 | −2.47 |
| RARA | Retinoic acid receptor, alpha | −2.44 |
With respect to endothelial dysfunction, DSP (desmoplakin) and SLC7A8 were up-regulated (Log2FC 1.44 and 1.27, respectively) in peripheral circulating cells of CAD+ patients compared to controls, whereas CDK5R2 (Log2FC −2.60) and CRHR1 (Log2FC −2.53) were down-regulated in CAD+ patients.
With respect to accumulation of circulating lipoproteins, the DAB1 gene was down-regulated in CAD+ patients (Log2FC −2.55) while the scavenger receptor MSR1 was the most up-regulated gene in CAD+ patients (Log2FC 1.62).
With respect to proliferation of the smooth muscle cell, LALBA (Log2FC −2.85), PDLIM4 (Log2FC −2.49), PRIMA 1 or MED24 (Log2FC −2.50) and NK2 (Log2FC −2.89) were down-regulated in CAD+ patients. Conversely, RAI2 (Log2FC 1.20) was up-regulated.
DiscussionOur results indicate that, in the patients in whom atherosclerosis, angiographically-assessed as absent, the RNA profile of leukocytes obtained from the systemic circulation (venous blood) was similar to that obtained from the coronary arteries. Conversely, in patients in whom atherosclerosis was present, there were significant differences in the gene expression profile of cells obtained from the coronary arteries. This is in agreement with the observations that the degree of inflammation is higher in the coronaries than in peripheral veins.6–8 Our results suggest that, in addition to the differences in gene expression extensively reported in the literature,2–5 the mere existence of regional differences in gene expression can be a distinctive trait of patients with established atherosclerosis. These data also support the concept of local predilective sites of inflammation and atherosclerosis.
Can these regional differences in RNA expression be explained by the differences in gender, systolic blood pressure and medications (such as aspirin and statin use) between the CAD+ and CAD− groups? A contribution of these factors cannot be ruled out, but the complete absence of differences between the systemic blood samples and those from the coronary arteries with respect to all biochemical parameters (particularly CRP and leucocyte count) seems to indicate the contrary.
The present study also shows that patients with CAD display highly significant differences in mRNA expression, defined as the means of all sites in nearly 2000 genes. From among these, we paid particular attention to the 20 genes most differentially expressed and which are mostly linked to the atherosclerosis process. From the literature, DSP, SLC7A8, CRHR1 and CDK5R2 are involved in endothelial dysfunction, the initial step in the atherosclerotic process; DSP encodes for desmoplakin and affects cell-to-cell junctions and endothelium permeability13; SLC7A8 interacts with ICAM-1 and increases cell adhesion14; CRHR1, which is associated with the production of adhesion molecules, was observed to be down-regulated in CAD thus, potentially, increasing permeability of circulating lipoproteins due to endothelial dysfunction15; CDK5R2 which phosphorylates NO synthase and is crucial in arterial vasodilatation, was also down-regulated in CAD.16
A key point in the progression of the atherosclerotic lesion is smooth cell proliferation, which, in part, resembles a tumour process. In the list of differentially expressed genes there were 4 tumour suppressors, 3 of which LALBA, PDLIM4 and MED24 were, indeed, down-regulated in patients with CAD. MED24 has been shown to decrease PPAR gamma.17 Additionally, RAI2, an inducer of proliferation, was up-regulated in CAD. The NK2 gene, which was down-regulated in CAD, binds to SMCs, depending on whether they are normal or neoplasic.18 All these genes are linked to endothelial dysfunction (DSP, SLC7A8, CRHR1 and CDK5R2) and smooth muscle cell proliferation (LALBA, PDLIM4, MED24, RAI2 and NK2). Of particular interest is that, among the macrophage scavenger receptor proteins (which are responsible for the uptake of lipoprotein remnants) the MSR1 gene was the most up-regulated in CAD, and was also one of the 4 genes differentially expressed consistently between groups (CAD+ vs. CAD−) and, consequently, for all sites separately as well.19 Other genes (RGPD1, GCC2 and FMO1) which were differentially expressed consistently between the two groups (CAD+ vs. CAD−) at all sites, have not had specific roles assigned, as yet, in atherogenesis.
Contrary to what we observed in the top 20 genes differentially expressed between CAD+ and CAD− patients (Table 3), the analyses of the genes differentially expressed between the systemic circulation (venous blood) and coronary arteries (arterial blood) from CAD+ patients, did not identify any block changes that could be associated with any known specific process or pathway within atherogenesis. Indeed, that there are significant differences in gene expression is highly relevant, and a critical review of the literature may provide further insight. For example, a number of studies5,20,21 have analysed differential gene expression in relation to atherosclerosis, and have described many genes differentially expressed that have some involvement in the process. These include cell migration and proliferation, endothelial dysfunction or signal transduction. However, there has not been high consistency of specific genes being identified in studies conducted to date.
This is a hypothesis-generating study and several limitations should be noted. Among the most important, the limited number of samples due to the difficulty in obtaining the specimens and the differences in treatments which we would anticipate to have influence on the comparison between cases and controls but little or no influence on the observed regional differences.
In summary, our study demonstrates that differential gene expression between venous blood sample and coronary artery blood samples might be an additional feature in established coronary artery disease. Whether differential expressions of these genes represent causes or effects of other processes occurring in these sites warrants further investigation.
Ethical disclosuresProtection of human and animal subjectsThe authors declare that the procedures followed were in accordance with the regulations of the responsible Clinical Research Ethics Committee and in accordance with those of the World Medical Association and the Helsinki Declaration.
Confidentiality of dataThe authors declare that they have followed the protocols of their work centre on the publication of patient data and that all the patients included in the study have received sufficient information and have given their informed consent in writing to participate in that study.
Right to privacy and informed consentThe authors have obtained the informed consent of the patients and/or subjects mentioned in the article. The author for correspondence is in possession of this document.
FundingThis study was supported by the Research Foundation Internal Specialties at Sint Franciscus Gasthuis, Rotterdam, The Netherlands.
Conflict of interestNone.