To compare the repetitive DNA patterns of human actinic keratoses and squamous cell carcinomas to determine the genetic alterations that are associated with malignant transformation.
INTRODUCTION:Cancer cells are prone to genomic instability, which is often due to DNA polymerase slippage during the replication of repetitive DNA and to mutations in the DNA repair genes. The progression of benign actinic keratoses to malignant squamous cell carcinomas has been proposed by several authors.
MATERIAL AND METHODS:Eight actinic keratoses and 24 squamous cell carcinomas (SCC), which were pair-matched to adjacent skin tissues and/or leucocytes, were studied. The presence of microsatellite instability (MSI) and the loss of heterozygosity (LOH) in chromosomes 6 and 9 were investigated using nine PCR primer pairs. Random Amplified Polymorphic DNA patterns were also evaluated using eight primers.
RESULTS:MSI was detected in two (D6S251, D9S50) of the eight actinic keratosis patients. Among the 8 patients who had squamous cell carcinoma-I and provided informative results, a single patient exhibited two LOH (D6S251, D9S287) and two instances of MSI (D9S180, D9S280). Two LOH and one example of MSI (D6S251) were detected in three out of the 10 patients with squamous cell carcinoma-II. Among the four patients with squamous cell carcinoma-III, one patient displayed three MSIs (D6S251, D6S252, and D9S180) and another patient exhibited an MSI (D9S280). The altered random amplified polymorphic DNA ranged from 70% actinic keratoses, 76% squamous cell carcinoma-I, and 90% squamous cell carcinoma-II, to 100% squamous cell carcinoma-III.
DISCUSSION:The increased levels of alterations in the microsatellites, particularly in D6S251, and the random amplified polymorphic DNA fingerprints were statistically significant in squamous cell carcinomas, compared with actinic keratoses.
CONCLUSION:The overall alterations that were observed in the repetitive DNA of actinic keratoses and squamous cell carcinomas indicate the presence of a spectrum of malignant progression.
Skin cancer is a multistep genetic disease that results from the accumulation of mutations in genes that are related to the cell cycle and the DNA repair systems at the root of genomic instability in tumor cells. Squamous cell carcinomas (SCCs) result from the malignant transformation of keratinocytes that reside in the squamous-cell layer of the epidermis, giving rise to primary tumors that metastasize in approximately 2.5% of the cases. SCCs and pre-neoplastic actinic keratoses (AKs) occur primarily on chronically sun-exposed sites of the body. UVB radiation (280-320 nm) is the prevailing cause of DNA damage of the skin, which may induce the progression of AKs to SCCs.1 Approximately 20% to 27% of SCCs arise in an AK or at a location that is nearby. Approximately 53,410 men and 60,440 women in Brazil were estimated to develop non-melanoma skin cancers as SCCs and basal-cell carcinoma (BCCs) in 2010.2
Approximately 90% of SCCs bear mutation signatures at dipyrimidic sites (C:G to T:A or CC:GG to TT:AA), particularly at the hotspots of the TP53 gene, which result in the inactivation of p53's functions in cell-cycle arrest at checkpoints, in DNA repair, or in the induction of apoptosis. An early occurrence of TP53 mutations has been associated with genomic instability and carcinogenic progression.1
Microsatellites are short, repeated sequences that are present at specific loci that frequently exhibit allelic polymorphisms (heterozygosity) and are useful as genetic markers for tumors. A gain or loss of internal repeats causes microsatellite instability (MSI), whereas the lack of an entire microsatellite results in loss of heterozygosity (LOH). MSI is associated with skin tumorigenesis3 and has also been shown to be an important factor in the cis-regulation of transcription.4
Alterations to several chromosomes (3p, 9p, 9q, 13q, 17p, and 17q) have been detected in SCCs. For example, the LOH at the 17p13.1 TP53 locus is an early event in tumor development and is consistent with the transition from AKs to noninvasive SCCs.5 Other microsatellite markers, which are located in the short (9p21) and long (9q22.3) arms of chromosome 9, exhibited LOH and/or MSI alterations in the AKs and SCCs. The number of alterations in 9p21 increased in the SCCs compared with the AKs; however, in both diseases, the incidence of MSI and LOH was similar. Other studies detected 14 LOHs and two MSIs in 27 SCCs for at least one microsatellite marker at 9p21 and detected mutations in the CDKN2A and CDKN2B tumor-suppressor genes. Specifically, the D9S50 exhibited six LOHs in 16 SCCs.6 Mortier et al.7 suggested that the malignant progression of AKs to SCCs is associated with the increased frequency of LOH at 9p21 and 17p in SCCs. A more recent study detected one LOH and five MSIs in 26 AK samples for at least one microsatellite marker at this locus.8 Our previous study on SCCs exhibited one LOH in four SCCs at the D9S50 and at 9p21.9 The mutations that were observed in the CDKN2A, CDKN2B, and TP53 genes and the reduced expression of the CDKN2A gene in the SCCs, compared with that in the AKs, reinforced the assumptions of many authors about the involvement of these genes in the progression of AKs to SCCs.
The genetic hallmark for the development of BCCs, another non-melanoma skin cancer, is the abrogation of the patched/sonic hedgehog pathway, which specifically involves the PTCH1 locus at 9q22.3-q31. Higher rates of LOH at 9q, 9p and 17p were found in AKs, rather than in invasive SCCs.10 D9S180 presented one MSI and one LOH out of 12 SCCs;11 an LOH in D9S180 was also detected in one out of two AKs and in two out of four SCCs. Regarding D9S287, a LOH was detected in one out of two AKs but was not detected in any SCCs.12
In contrast to the microsatellite analysis method, the RAPD (random amplified polymorphic DNA) method detects genetic alterations in the entire genome using single primers that are composed of ten arbitrary oligomers and give rise to personal fingerprints.13 An analysis of 30 head and neck SCCs with 10 primers detected alterations in 100% of the samples with at least one RAPD primer. There was a significant correlation between the tumor stage and the frequency of alteration.14
Our previous study on SCCs and AKs indicated the presence of altered RAPD fingerprints with OPA-2 and OPB-15 primers in 100% of the samples, compared with normal tissues.9
The results of this previous study encouraged us to investigate the genomic instability of repetitive DNA in an enlarged cohort of AKs, SCCs, and matched normal tissues using eight primers for RAPD and nine for microsatellites.
MATERIAL AND METHODSTissues and DNA extractionThe DNA was extracted from eight AKs and 24 SCC lesions (10 SCC-I, 10 SCC-II, and four SCC-III). Supplementary Table 1s provides the following characteristics of the studied patients: the identification number attributed to each patient in the laboratory list, age, sex, body site of the tumor, and associated skin tumors. The tumors and paired adjacent normal tissues or blood leucocytes were obtained from patients who were diagnosed and treated in the Outpatient Clinics of the Department of Dermatology, Faculdade de Medicina da Universidade de Sao Paulo. These patients signed an informed consent form, and the research was approved by the Ethical Committee of the Institution (CAPPesq). The procedure for extracting the DNA from the tumors included biopulverization of the tumor tissues, digestion in TES buffer (10 mM Tris pH 8.0, 10 mM EDTA, 0.6% SDS) that contained proteinase K 100 ug/ml at 50 °C for 3-4 h, DNA extraction with phenol/chloroform/isoamyl alcohol, and ethanol precipitation. This procedure was performed according to standard protocols. Leucocytes were included in the present study to evaluate the possible differences between the repetitive patterns that were present in this type of normal tissue and the normal skin that surrounded each tumor. Following the lysis of the red blood cells (in a 10x buffer that consisted of 1550 mM NH4Cl, 100 mM KHCO3, and 10 mM EDTA), the leucocytes' DNA was extracted as described above. The malignancy grading of the SCCs was defined, according to the Broders classification,15 as either well differentiated (SCC-I), moderately differentiated (SCC-II), or poorly differentiated (SCC-III).
The RAPDs and microsatellites studied, including the primer sequences, PCR annealing temperatures, chromosomal locations and polymorphism ranges in base pairs (bp).*
| Markers | Primers sequences | Annealing t° | Chromosome |
|---|---|---|---|
| D6S251 | 5′- TTCCTAACCAGGTTTCAATG -3′ | 51°C | 6q14-16 |
| (GT)n | 5′- ATATTTTTAAAGTAAGTTGCAC -3′ | 144-162 bp* | |
| D6S252 | 5′- ATGGCTCAGGATTCACATTG -3′ | 53°C | 6q14-16 |
| (GT)n | 5′- TGAAAGGAAAGTCCTGCTTC -3′ | 154-166 bp* | |
| D9S15 | 5′- TAAAGATTGGGAGTCAAGTA -3′ | 55°C | 9q13-21 |
| (CA)n | 5′- TTCACTTGATGGTGGTAATC -3′ | 195-209 bp* | |
| D9S50 | 5′- GATCCTTTTCATCTTCTGAC -3′ | 52°C | 9p21 |
| (CAGT)n | 5′- GAGGGACGGAGCAACTGAT -3′ | 76-100 bp* | |
| D9S52 | 5′- GCTAGAGGGAGGTTTAATTG -3′ | 54°C | 9p21 |
| (ATTT)n | 5′- AATTAGCCAGGTGTTGTGGT -3′ | 213-229 bp* | |
| D9S180 | 5′- CAGTGGTTTGGAATCGAACC -3′ | 55°C | 9q22.3 |
| (CA)n | 5′- AGCTATTTTTGGGGGCTGAG -3′ | 220-230 bp* | |
| D9S196 | 5′- GGGATTACACCTCAAAACCA -3′ | 55°C | 9q22.3 |
| (CA)n | 5′- ACCACACTGCGGGACTT -3′ | 254-260 bp* | |
| D9S280 | 5′- TTTCGCTTCCCACCCA -3′ | 55°C | 9q22.3 |
| (CA)n | 5′- CACGCCACTGATCTAGGCT -3′ | 140-160 bp* | |
| D9S287 | 5′- GGATGCTCCTCACGC -3′ | 55°C | 9q22.3 |
| (CA)n | 5′- ACCACTACATTGTTCAAGGG -3′ | 168-180 bp* | |
| OPA-2 | 5′- TGCCGAGCTG -3′ | 34°C | |
| OPA-7 | 5′- GAAACGGGTG -3′ | 35°C | Unspecific |
| OPA-13 | 5′- CAGCACCCAC -3′ | 34°C | fingerprints, |
| OPA-17 | 5′- GACCGCTTGT -3′ | 32°C | concerning the |
| OPB-8 | 5′- GTCCACACGG -3′ | 34°C | whole genome; |
| OPB-11 | 5′- GTAGACCCGT -3′ | 32°C | sizes of the |
| OPB-13 | 5′- TTCCCCCGCT -3′ | 32°C | variable bands |
| OPB-19 | 5′- ACCCCCGAAG -3′ | 34°C |
Nine microsatellite primers were used to amplify the DNA sequences that were located in the human chromosomes 6 and 9 by PCR to verify the genomic instability that was related to these chromosomal sites. The following microsatellites were studied: D6S251 and D6S252, which are located at 6q14-16; D9S15, which is located at 9q21.11, a highly repetitive chromosomal region; D9S50 and D9S52, which are located at 9p21, near the CDKN2A and CDKN2B genes; D9S180, D9S280, and D9S287, which are located downstream to the PTCH1 gene at 9q22.3; and D9S196, which is located upstream of the PTCH1 gene. The PCR analyses were conducted in a final volume of 50 μl, containing 10 pM of each primer, 100 ng DNA, 10X PCR buffer, 1.5 mM MgCl2, 0.2 mM dNTP mix, and 1 unit Taq DNA polymerase (LGC Biotechnology). The thermocycling conditions consisted of 40 cycles of 1 min at 96 °C, with variable annealing temperatures (Table 1) and an extension of 1 min at 72 °C. The PCR products were resolved and visualized on a silver-stained 7% polyacrylamide gel. When the signal intensity of the band from the tumor DNA was reduced by 50% or greater, it was considered to be indicative of an LOH. Bands with decreased or increased sizes were defined as examples of an MSI. The images were captured using the Kodak 1D 3.6 Scientific Imaging System® under a UV light. The number of samples that were studied for each microsatellite and the corresponding histological grades are depicted in Supplementary Table 2s.
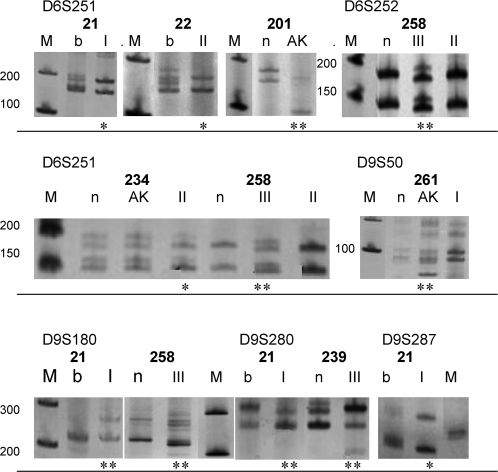
The patients who carried MSI or LOH mutations were indicated for each microsatellite, according to their laboratory list number. The bottom row indicates the total number of alterations found in all of the patients with informative results. (*) The total alterations of D6S251 were significantly greater in the SCC (I+II+III) compared with the AK lesions (p = 0.0398).
| D6S251 | D6S252 | D9S50 | D9S180 | D9S280 | D9S287 | ||
|---|---|---|---|---|---|---|---|
| LOH | MSI | MSI | MSI | MSI | MSI | LOH | |
| AK 1/8 | 201 | 261 | |||||
| SCC-I 1/8 | 21 | 21 | 21 | 21 | |||
| SCC-II 3/10 | 22, 234 | 258 | |||||
| SCC-III 2/4 | 258 | 258 | 258 | 239 | |||
| 6/30* | 1/33 | 1/31 | 2/30 | 2/26 | 1/31 | ||
The RAPD primers OPA-2, OPA-7, OPA-13, OPA-17, OPB-8, OPB-11, OPB-13, and OPB-19 (Operon Technologies, Alameda, CA) were used to evaluate the whole-genome instability of the cancer cells, compared with normal tissues. The PCR analyses were conducted in a final volume of 30 μl using 50 ng DNA, 10X PCR buffer, 2.5 mM MgCl2, 330 μM dNTP mix, and 1 unit Taq DNA polymerase (LCG Biotechnology). The OPA or OPB primers (10 pM) were added to the reaction mix, and the reaction included 45 cycles consisting of 1 min for denaturation at 94 °C, 1 min at a variable annealing temperature (Table 1), 1 min of extension at 72 °C, and a final extension of 5 min at 72 °C. The PCR-amplified fragments were visualized on an ethidium bromide-stained 1.5% agarose gel and were photographed under a UV light. The bands that did not match the normal pattern and/or presented an enhanced fluorescence signal following the ethidium bromide staining were defined as having a “band gain.” Any missing or weakly stained bands in the tumor pattern were counted as having a “band loss.” The total number of the tumors/controls that were studied with each RAPD primer and the corresponding histological types are presented with the results.
Statistical analysisThe number of alterations in the repetitive DNA patterns of the tumors was compared with that of the controls using the Fisher test, which was included in the GraphPad InStat 3 Program. The differences were considered to be significant at p≤0.05.
RESULTSMicrosatellite analysisTable 2 identifies the patients who presented LOHs and/or MSIs and shows the total number of alterations in each microsatellite that was detected in all of the samples, which are illustrated in Fig. 1. Interestingly, two MSIs and two LOHs were detected in a single SSC-I from patient 21, who also exhibited one BCC. One and three MSIs were found in one SSC-II and one SSC-III, respectively, with both tumors being derived from patient 258, who also exhibited multiple BCCs.
The D6S251, D6S252, D9S50, D9S180, D9S280, and D9S287 microsatellites were visualized on a silver nitrate-stained polyacrylamide gel. Patient identification (bold numbers). Leucocytes (b), normal tissue (n), actinic keratosis (AK), and squamous cell carcinoma (SCC) grades I, II, and III. (*) LOH, (**) MSI, (M) Molecular marker (bp).
The AK or SCC samples did not exhibit alterations in D9S15, D9S52, or D9S196. Furthermore, no microsatellite alterations were observed in the associated BCC that was present in patients 261, 274, and 275.
The patients bearing SCCs of grades I, II, and III exhibited a greater number of LOHs and/or MSIs than did those bearing AKs, with two alterations in eight AKs, four alterations (in a single tumor) in 10 SCC-Is, three alterations in 10 SCC-IIs, and four alterations (in two tumors) in four SCC-III being observed. However, the results were not significant when the differentiation grades of the SCCs were considered. D6S251 was the most altered microsatellite (p = 0.0398), exhibiting six alterations in 30 samples (20%).
RAPD ANALYSISThe number of RAPD bands in the SCCs tended to be greater than that of the AKs; however, these differences were not statistically significant (data not shown).
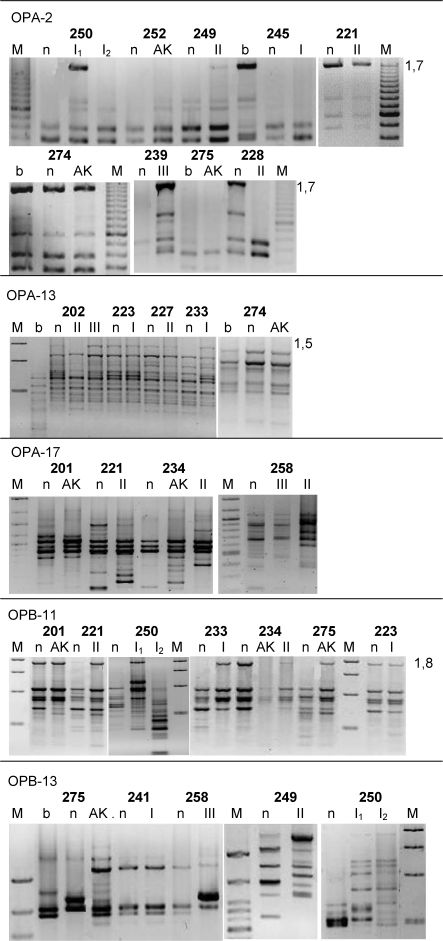
In general, the OPA-2 and OPA-13 patterns were highly conserved, whereas the other primers resulted in more variable fingerprints. Two AKs, one SCC-I, and one SCC-II were not altered compared to the corresponding control. Various lesions that were present in the same patient had distinct patterns, indicating that unrelated genetic changes were present among these tumors. The AKs' patterns were more similar to those of the controls than to those of the SCCs that were present in the same patient, confirming the presence of increased alterations during the progression of malignancy. Figure 2 and Supplementary Figure 1s show the other representative RAPD patterns. Several differences in patterns that were observed between the leucocytes and the adjacent skin tissues (Fig. 2) may be accounted for by the normal genomic rearrangements that result in the various T-cell lineages that are present in the blood.
The RAPD patterns that were obtained using primers OPA-2, OPA-13, OPA-17, and OPB-11 were visualized on an ethidium bromide-stained gel. Several bands that were altered in the tumors are indicated (kb). The abbreviations are as indicated in Fig. 1.
Table 3 lists the absolute numbers and percentages of tumors with altered fingerprints (as compared with normal tissues), taking into consideration the total number of samples that were studied with each primer. The mean value ± the standard deviation of the altered fingerprints was 0.7075±0.08992 for AK, 0.7588±0.04291 for SCC-I, 0.9063±0.04625 for SCC-II, and 0.9950±0.001890 for SCC-III. The statistical analysis (t-tests and non-parametric tests) indicated significant results for AK versus SCC-III (p = 0.0065), SCC-I versus SCC-II (p = 0.0347), and SCC-I versus SCC-III (p<0.0001). Two of the associated BCCs that were studied were found to be altered when they were investigated with OPA-7, OPA-17 and OPB-8 primers, but not when they were investigated with OPA-2 primers (not shown).
The RAPD patterns were significantly altered in the tumors compared with the normal tissues (%): AK x SCCIII (p = 0.0065), SCCI x SCCII (p = 0.0347), and SCCI x SCCIII (p<0.0001).
| OPA | OPB | Mean % | |||||||
|---|---|---|---|---|---|---|---|---|---|
| 2 | 7 | 13 | 17 | 8 | 11 | 13 | 19 | ||
| AK | 3/7 42% | 6/7 85% | 5/6 83% | 1/3 33% | 4/7 57% | 4/6 66% | 5/5 100% | 7/7 100% | 70% |
| SCC I | 6/7 85% | 6/7 85% | 4/7 57% | 4/5 80% | 4/7 57% | 5/6 83% | 3/4 75% | 6/7 85% | 82% |
| SCC II | 5/8 62% | 7/8 87% | 8/9 88% | 6/6 100% | 8/9 88% | 9/9 100% | 8/8 100% | 8/8 100% | 90% |
| SCC III | 4/4 100% | 4/4 100% | 2/2 100% | 4/4 100% | 2/2 100% | 4/4 100% | 4/4 100% | 4/4 100% | 100% |
Repetitive DNA patterns were investigated to determine the possible markers that were associated with the malignant transformation of AKs to SCCs.
Among the nine microsatellites that were studied, D6S251 was the most altered in the SCC lesions, followed by D9S180 and D9S280 (Table 2). D6S252 demonstrated only one MSI in one SCC-III. Our previous BCC studies also identified a greater number of alterations in D6S251 than in D6S252. One study of BCCs found a 60% MSI and a 10% LOH in D6S251 and only a 10% MSI in D6S252.16 Another study detected a 7.6% MSI and a 15% LOH in D6S251 in the high-risk BCCs but detected no alterations in D6S252.17 On the contrary, D6S252 exhibited a 30% MSI, and D6S251 exhibited only a 9% LOH in an MM study (Lozano L.F., Master Thesis, 2009).18
Reports of altered D6S251 and D6S252 sequences in AK and SCC genomes were not found in the literature, and no genes related to SCC development have been observed adjacent to these microsatellites. However, abnormalities in the 6q12-q21 chromosomal region have been extensively observed in several cancer types, particularly in MM. Cytogenetic studies have revealed complex rearrangements and deletions at 6q and have revealed an LOH at the molecular level, suggesting the existence of a putative suppressor gene that may be found farther down in this region.
For chromosomal region 9p, our previous study found an LOH at D9S50 in one of the four SCCs.9 However, in the present, large study of 22 SCCs, no alterations in this microsatellite were detected, and only one MSI in seven AKs was observed. Other studies detected an LOH in the 9p21 region in SCCs and AKs,7 which would have altered the expression of the tumor-suppressor genes CDKN2A and CDKN2B. These genes are involved in the progression of AKs to SCCs.19 These genes did not seem to be related to the malignant progression of our samples.
Mutations in the PTCH1 gene at 9q were associated with the development of >70% of the BCCs and with the development of SCCs in individuals with histories of multiple BCCs.20 Alterations in the microsatellites that were adjacent to PTCH1 may have affected the expression of the PTCH1 protein in the SCCs but not in the AKs and BCCs, which were not altered in these individuals.
Several authors have considered AK to be an initial step of a spectrum of disease that leads to SCC; however, as already mentioned, the data are controversial. Japanese patients exhibited an LOH of several microsatellites in 19% of the AKs and in 7% of the SCCs, but no MSIs were observed. According to the authors of that study, these results were not indicative of tumor progression from AKs to SCCs, and the molecular pathogenesis in skin tumors may be race- and geography-dependent.21 Other studies have also observed LOHs at a higher frequency in AKs than in SCCs, suggesting that AKs are not precursors to SCCs.5,9 Conversely, the AK samples that were studied in the present work did not exhibit an LOH but observed two MSIs, whereas the SCCs presented four LOHs and eight MSIs. Padilla et al.22 have recently identified common genes whose expression was up- or downregulated in AKs and SCCs. However, these genetic characteristics were not differentiated between the AKs and SCCs.
In addition to the microsatellite analysis, three RAPD primers (OPA-2, OPA-7 and OPB-8) revealed 86% and 95% alterations in the AKs and SCCs, respectively. Our previous data indicated that 100% alterations occurred with the primers OPA-2 and OPB-15 in all of the skin-lesion types, including AKs, SCCs, BCCs, MM, and MN (melanocytic nevus).9
Overall, these results demonstrated highly conserved RAPD patterns in SCCs from various patients, and especially in the case of AKs (cf. Fig. 2, OPA-2 and OPA-13, and Supplementary Figure 1s). The total number of alterations in the RAPD patterns was, in all cases, greater in the SCCs than AKs when compared with the patterns that were observed in the corresponding normal tissues (p<0.01). A significant correlation between the SCCs' malignancy grading and the frequency of the RAPD alterations was observed. Maeda et al. described the genomic instability using at least one RAPD primer in 100% of the head and neck SCCs and also observed a significant correlation with the tumor differentiation degree.14 A similar correlation was also observed in hepatocellular carcinomas23 and breast cancers.24
Because the differences between the numbers of gained and lost bands in the fingerprint profiles were not statistically significant, the putative rearrangements of the tumor genomes must have been balanced, without preferential increases or losses of genetic material. The type of genomic rearrangements that result in altered tumor fingerprints should be investigated by isolating, cloning, and sequencing the particular bands that were missing or were newly found in the DNA patterns of several patients. Further analyses of these DNA fragments may yield significant information regarding the molecular mechanisms of SCC development and may eventually reveal new biomarkers. The overall alterations that were found in the repetitive DNA patterns in the present study reinforce the assumption of a spectrum of malignant progression from AKs to SCCs.
We are indebted to Marcos A. R. Martinez, MD; Maria Cristina Messina, MD, MS; Daniela C. C. Santos, MS; Adriana M. Orimoto, MS; and Alberto O. Cardoso, MD, MS for providing some of the control and tumor samples or DNA. This work was supported by the FAPESP.