Incidence of childhood-onset type 1 diabetes mellitus in the Canary Islands is the highest reported so far in Spain, and among the highest worldwide. The HLA region accounts for approximately half the genetic risk of type 1 diabetes. Our aim was to assess distribution of high-risk and protective HLA haplotypes in the Canarian families included in the T1DGC, as compared to the rest of Spain.
MethodsThe T1DGC study, an international project to study the genetics and pathogenesis of type 1 diabetes, enrolled more than 3000 families with type 1 diabetes worldwide. Spain provided 149 of these families, of whom 42 were from Tenerife and Gran Canaria. HLA was genotyped centrally using a PCR-based, sequence-specific oligonucleotide probe system. Haplotypes were reconstructed using the deterministic algorithm alleHap in the R programming environment. Based on prior T1DGC results in Caucasian population, haplotypes DRB1*0405-DQA1*0301-DQB1*0302, DRB1*0401-DQA1*0301-DQB1*0302, DRB1*0301-DQA1*0501-DQB1*0201, DRB1*0402-DQA1*0301-DQB1*0302 and DRB1*0404-DQA1*0301-DQB1*0302 were considered high-risk. DRB1*0701-DQA1*0201-DQB1*0303, DRB1*1401-DQA1*0101-DQB1*0503, DRB1*1501-DQA1*0102-DQB1*0602, DRB1*1101-DQA1*0501-DQB1*0301, DRB1*1104-DQA1*0501-DQB1*0301, DRB1*1303-DQA1*0501-DQB1*0301, DRB1*1301-DQA1*0103-DQB1*0603 and DRB1*0403-DQA1*0301-DQB1*0302 were considered protective. The distribution of protective, high-risk, and other haplotypes in the (first two) affected siblings and unaffected parents from Canarian and non-Canarian Spanish families was compared (Chi-square test).
ResultsNo significant differences were found between the regions in distribution of the HLA haplotypes in the affected siblings or in the non-affected parents.
ConclusionsThe high incidence of childhood-onset type 1 diabetes in the Canarian population does not appear to be explained by a greater prevalence of high-risk class II HLA haplotypes in families with the disease. However, sample size limits the differences that can be detected in this study.
La incidencia de diabetes tipo 1 infantil en Canarias es la más alta descrita hasta el momento en España y una de las mayores a nivel mundial. La región HLA explica aproximadamente el 50% del riesgo genético de la diabetes tipo 1. Nuestro objetivo fue comparar la frecuencia de haplotipos de HLA de riesgo y protectores en familias españolas canarias y peninsulares incluidas en el T1DGC.
MétodosEl T1DGC es un proyecto internacional que estudia la genética y patogenia de la diabetes tipo 1, para el que fueron inluidas más de 3000 familias con la enfermedad. Un total de 149 familias provenían de España, y 42 de ellas, de Tenerife y Gran Canaria. El HLA fue genotipado en un laboratorio central, utilizando un método basado en PCR y sondas específicas de secuencia. Los haplotipos fueron reconstruidos utilizando el algoritmo determinista alleHap en el entorno de programación R. En base a los resultados previos del T1DGC en población caucásica, los haplotipos DRB1*0405-DQA1*0301-DQB1*0302, DRB1*0401-DQA1*0301-DQB1*0302, DRB1*0301-DQA1*0501-DQB1*0201, DRB1*0402-DQA1*0301-DQB1*0302 y DRB1*0404-DQA1*0301-DQB1*0302 fueron definidos como de alto riesgo. DRB1*0701-DQA1*0201-DQB1*0303, DRB1*1401-DQA1*0101-DQB1*0503, DRB1*1501-DQA1*0102-DQB1*0602, DRB1*1101-DQA1*0501-DQB1*0301, DRB1*1104-DQA1*0501-DQB1*0301, DRB1*1303-DQA1*0501-DQB1*0301, DRB1*1301-DQA1*0103-DQB1*0603 y DRB1*0403-DQA1*0301-DQB1*0302 fueron considerados protectores. La distribución de haplotipos de riesgo, protectores y otros en los (dos primeros) hermanos afectos y en los padres no afectos fue comparada entre las familias canarias y no canarias (chi cuadrado).
ResultadosNo se encontraron diferencias significativas en la distribución de haplotipos HLA entre las regiones estudiadas, ni en los hermanos afectos ni en los padres no afectos.
ConclusionesLa alta incidencia de la enfermedad en la población canaria no parece ser explicada por una mayor prevalencia de haplotipos de HLA de clase II de riesgo en los casos con agregación familiar, aunque el tamaño de la muestra limita las diferencias detectables en este estudio.
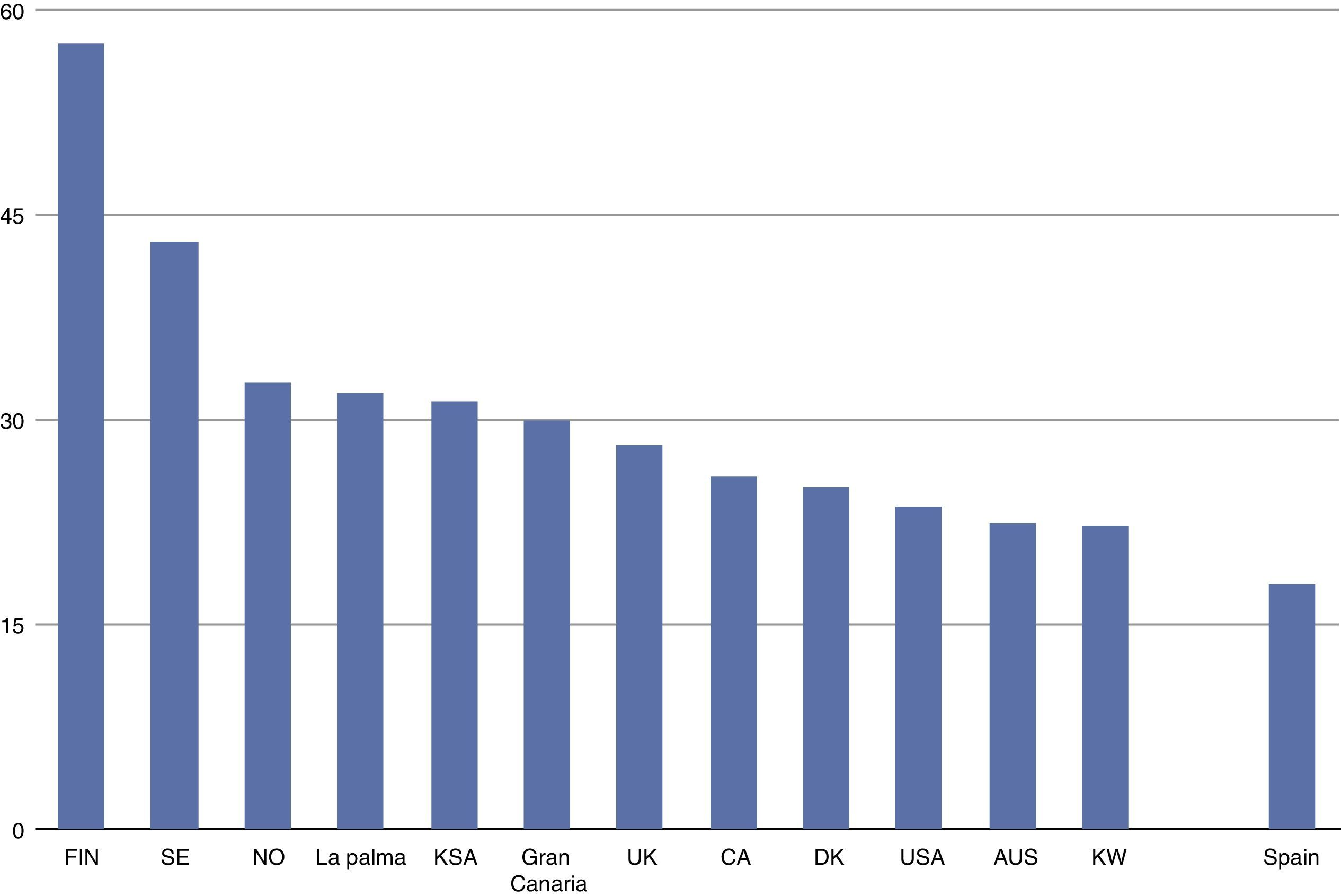
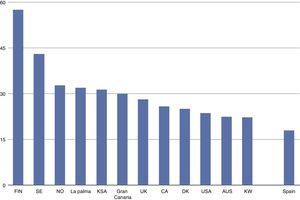
The incidence of childhood-onset type 1 diabetes in the Canary Islands is the highest described so far in Spain1–4 and one of the highest worldwide5 (see Fig. 1), though no genetic or environmental explanation has yet been found.
Incidence of childhood-onset type 1 diabetes (cases/100,000 inhabitants, based on 1, 3–5). T1D: Type 1 diabetes; FIN: Finland; SE: Sweden; NO: Norway; KSA: Kingdom of Saudi Arabia; UK: United Kingdom; CA: Canada; DK: Denmark; USA: United States of America; AUS: Australia; KW: Kuwait.
Human Leucocyte Antigen (HLA) accounts for about fifty percent of the genetic risk of type 1 diabetes6 and we hypothesised that a difference in the prevalence of high-risk or protective alleles might be a possible explanation for this high incidence in the Islands.
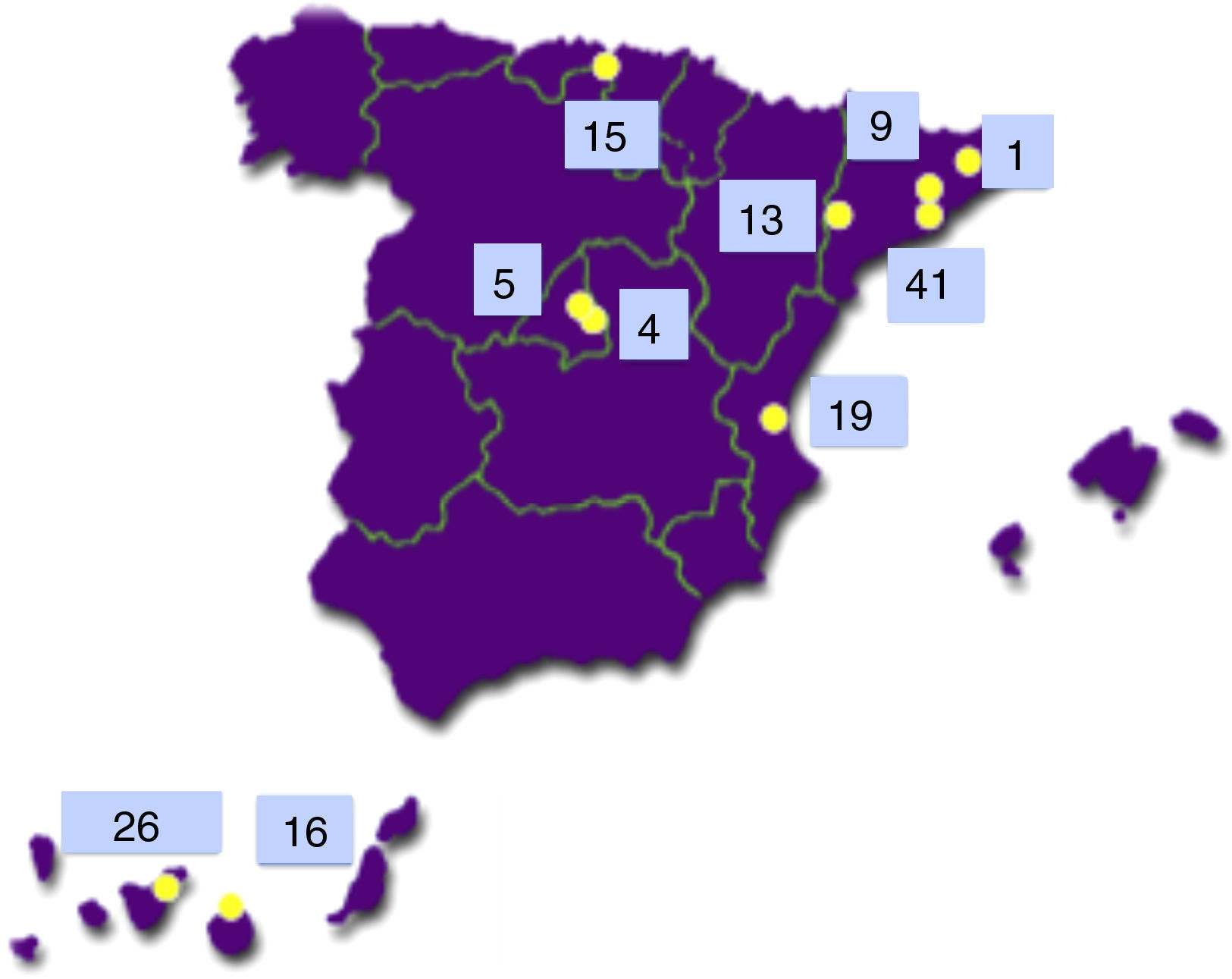
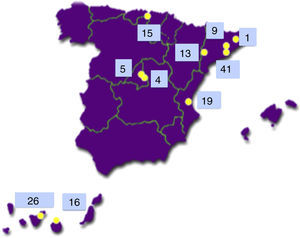
The Type 1 Diabetes Genetics Consortium (T1DGC) is an international endeavour to study the genetics and pathogenesis of type 1 diabetes.7 Of the more than 3000 families with type 1 diabetes included worldwide, 149 came from Spain and 42 of them (28.2%) were from the Canary Islands Tenerife (26) and Gran Canaria (16)8 (see Fig. 2).
Our aim was to assess high-risk and protective, class II (DRB1-DQA1-DQB1) HLA haplotype distribution in the Canarian families, compared with those from the rest of Spain, included in the T1DGC.
MethodsFamilies were included in the study if at least two siblings had type 1 diabetes. Both affected and unaffected siblings were invited to participate, as well as their parents. The study was approved by the centers’ ethics committees and participants signed a written, informed consent form. Clinical information was collected by means of standardised questionnaires delivered at each of the participating centres.9 Blood samples were obtained and HLA was genotyped centrally in Malmo¿, Sweden, using a PCR-based, sequence-specific oligonucleotide probe system and alleles were read with specific software (SCORE).10
The HLA haplotypes were constructed using the R package alleHap, a deterministic algorithm for imputing missing genetic data and reconstructing haplotypes from pedigree databases.11 Risk and protective class II HLA haplotypes were defined, based on previous T1DGC results in Caucasian populations.10 Briefly, haplotypes DRB1*0405-DQA1*0301-DQB1*0302, DRB1*0401-DQA1*0301-DQB1*0302, DRB1*0301-DQA1*0501-DQB1*0201, DRB1*0402-DQA1*0301-DQB1*0302 and DRB1*0404-DQA1*0301-DQB1*0302 were considered high-risk, whereas DRB1*0701-DQA1*0201-DQB1*0303, DRB1*1401-DQA1*0101-DQB1*0503, DRB1*1501-DQA1*0102-DQB1*0602, DRB1*1101-DQA1*0501-DQB1*0301, DRB1*1104-DQA1*0501-DQB1*0301, DRB1*1303-DQA1*0501-DQB1*0301, DRB1*1301-DQA1*0103-DQB1*0603 and DRB1*0403-DQA1*0301-DQB1*0302 were considered protective. All haplotypes not included in the mentioned categories were defined as “other”.
In the case of incomplete haplotype reconstruction, if only one option of complete haplotype was possible according to the rest of the database, then the appropriate, complete haplotype would be inferred and classified. For example, in the case of the incomplete DRB1*0301-DQA1*?-DQB1*0201, the high-risk DRB1*0301-DQA1*0501-DQB1*0201 haplotype would be inferred. If several alternatives were possible, albeit in the same category, then the haplotype was also classified. For example, DRB1*1303-DQA1*?-DQB1*0402 could be translated into different possibilities, although none of them classified as high-risk or protective. Thus, it would be classified as “other”. Finally, if several alternatives were possible, in different categories, the haplotype would be considered “lost” and would not be included in the analysis. For example, DRB1*?-DQA1*0301-DQB1*0302, could be both high-risk or protective and would thus be declared missing.
The distribution of protective, high-risk and other haplotypes was compared in the first two affected siblings per family and in the unaffected parents in the Canarian and non-Canarian Spanish participants, using chi-squared. Since the majority of the non-Canarian families were recruited in Catalonia, a separate analysis was also performed, comparing the Canarian and Catalonian families.
ResultsFully or partially complete (one allele missing), unambiguous DRB1-DQA1-DQB1 HLA haplotypes were obtained in 144 Canarian (74 siblings with type 1 diabetes and 70 non-diabetic parents) and 301 non-Canarian subjects (162 siblings with type 1 diabetes and 139 non-diabetic parents). This led to 456 complete unambiguous and 16 partially complete haplotypes among the affected siblings and 411 and 7, respectively, among the unaffected parents, that could be classified according to their risk. There were no significant differences between regions in the distribution of the haplotypes into their risk categories for the siblings or for the non-affected parents (see Table 1). Table 2 displays the frequency of selected, specific, complete, unambiguous haplotypes in the affected siblings and in the parents. The comparison between the Canarian and Catalonian families did not show any significant differences, either (data not shown).
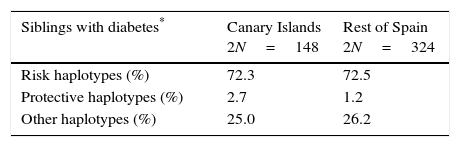
High-risk and protective, class II HLA haplotypes in siblings with type 1 diabetes and in their non-affected parents (%2N).
| Siblings with diabetes* | Canary Islands 2N=148 | Rest of Spain 2N=324 |
|---|---|---|
| Risk haplotypes (%) | 72.3 | 72.5 |
| Protective haplotypes (%) | 2.7 | 1.2 |
| Other haplotypes (%) | 25.0 | 26.2 |
| Parents without diabetes** | Canary Islands 2N=140 | Rest of Spain 2N=278 |
|---|---|---|
| Risk haplotypes (%) | 47.9 | 51.4 |
| Protective haplotypes (%) | 9.2 | 13.7 |
| Other haplotypes (%) | 42.9 | 34.9 |
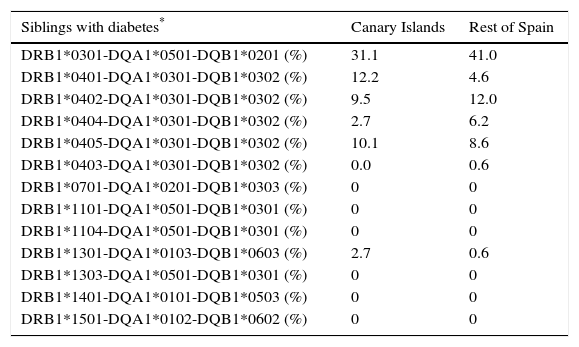
Distribution of some specific, unequivocal haplotypes.
| Siblings with diabetes* | Canary Islands | Rest of Spain |
|---|---|---|
| DRB1*0301-DQA1*0501-DQB1*0201 (%) | 31.1 | 41.0 |
| DRB1*0401-DQA1*0301-DQB1*0302 (%) | 12.2 | 4.6 |
| DRB1*0402-DQA1*0301-DQB1*0302 (%) | 9.5 | 12.0 |
| DRB1*0404-DQA1*0301-DQB1*0302 (%) | 2.7 | 6.2 |
| DRB1*0405-DQA1*0301-DQB1*0302 (%) | 10.1 | 8.6 |
| DRB1*0403-DQA1*0301-DQB1*0302 (%) | 0.0 | 0.6 |
| DRB1*0701-DQA1*0201-DQB1*0303 (%) | 0 | 0 |
| DRB1*1101-DQA1*0501-DQB1*0301 (%) | 0 | 0 |
| DRB1*1104-DQA1*0501-DQB1*0301 (%) | 0 | 0 |
| DRB1*1301-DQA1*0103-DQB1*0603 (%) | 2.7 | 0.6 |
| DRB1*1303-DQA1*0501-DQB1*0301 (%) | 0 | 0 |
| DRB1*1401-DQA1*0101-DQB1*0503 (%) | 0 | 0 |
| DRB1*1501-DQA1*0102-DQB1*0602 (%) | 0 | 0 |
| Parents without diabetes** | Canary Islands | Rest of Spain |
|---|---|---|
| DRB1*0301-DQA1*0501-DQB1*0201 (%) | 19.3 | 30.0 |
| DRB1*0401-DQA1*0301-DQB1*0302 (%) | 7.1 | 2.9 |
| DRB1*0402-DQA1*0301-DQB1*0302 (%) | 7.9 | 8.3 |
| DRB1*0404-DQA1*0301-DQB1*0302 (%) | 2.9 | 5.0 |
| DRB1*0405-DQA1*0301-DQB1*0302 (%) | 7.6 | 5.4 |
| DRB1*0403-DQA1*0301-DQB1*0302 (%) | 0.7 | 1.8 |
| DRB1*0701-DQA1*0201-DQB1*0303 (%) | 0 | 0 |
| DRB1*1101-DQA1*0501-DQB1*0301 (%) | 0 | 1.1 |
| DRB1*1104-DQA1*0501-DQB1*0301 (%) | 0 | 0.7 |
| DRB1*1301-DQA1*0103-DQB1*0603 (%) | 5.7 | 3.6 |
| DRB1*1303-DQA1*0501-DQB1*0301 (%) | 0.7 | 1.1 |
| DRB1*1401-DQA1*0101-DQB1*0503 (%) | 0.7 | 0 |
| DRB1*1501-DQA1*0102-DQB1*0602 (%) | 0.7 | 5.0 |
Note: The high risk and protective haplotypes may not exactly add to the corresponding column in Table 1 due to partially complete haplotypes that could be classified into high risk, (10 in the siblings and 4 in the parents) protective (1 in the parents) or other (6 in the siblings and 2 in the parents).
According to this family-based study, the high incidence of childhood-onset type 1 diabetes in the Canarian population does not seem to be explained by higher-risk class II HLA haplotypes.
The strengths of this study include its family-based nature and the use of standardised methods for data and sample collection and processing, which allowed for haplotype imputation and direct comparison.
Previous studies performed in Spain show a high frequency of high-risk HLA haplotypes in patients with type 1 diabetes, comparable to other populations. A higher frequency and higher risk associated with DR3-DQ2 (compared with DR4-DQ8) has been found in mainland Spain in previous reports.12–14 Furthermore, populations-specific, extended DR3 HLA haplotypes have been identified.15 In the present study, DR3 was the most frequent class II HLA in peninsular Spain, but not in the Canary Islands. In a recent study performed in Gran Canaria in children with type 1 diabetes, more than 95% of the participants showed at least one DQ2 or DQ8 allele (which are often assumed to be part of high-risk DR3-DQ2 and DR4-DQ8 haplotypes, respectively).3 In that study 40% of alleles were DQ2 and 36% were DQ8,3 which does not fully agree with the presently described results. Indeed, this family-based population, with at least two affected siblings per family, may not necessarily be representative of the more frequent, sporadic form of type 1 diabetes. Furthermore, the families included in the T1DGC need not be strictly representative of all families with type 1 diabetes. Finally, a type II error due to small sample size cannot be ruled out. In the present study, differences between groups ranged between 1.2 and 1.5% in the case of the affected siblings, and between 3.5 and 8% in the case of the non-affected parents. For the study to have an 80% power to detect a 1.5% difference, the sample should have included 8000 participants per group (8000 Canarian and 8000 non-Canarian) and, to detect a 3.5% difference, we would still need about 1600 individuals per group.
Although the Canarian population is mostly of Iberian-European descent,16 North African genetic markers are also present.17 In fact, studies performed in the North of Africa also identify DR3 and DR4 as the main high-risk alleles associated with type 1 diabetes.18,19 More extensive haplotyping might allow for the identification of population-specific haplotypes, as in other Spanish regions. On the other hand, different genotyping methods, with varying resolutions, limit the comparisons that can be made among studies. Furthermore, haplotype imputation is often not possible, especially in case-control studies, where parents might not be available for genotyping.
HLA accounts for most, albeit not all the genetic risk of type 1 diabetes. About 40 additional genetic loci have been associated with moderate disease susceptibility, although only variants in INS, PTPN22, CTLA4, and IL2RA are associated with OR above 1.1.6 Although they have not been assessed in the current analysis, they should be available for future studies.
Several environmental factors have also been associated with the risk of type 1 diabetes, including virus infections, early dietary patterns and vitamin D.6 These and/or other environmental or genetic factors should be sought that may account for the high incidence of childhood-onset type 1 diabetes in the Canary Islands.
In conclusion, despite a higher incidence of childhood-onset type 1 diabetes in the Canary Islands, our family-based results do not show an increased prevalence of high-risk HLA haplotypes when compared with mainland Spain.
FundingThis research uses resources provided by the Type 1 Diabetes Genetics Consortium (T1DGC), the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK, U01 DK062418), the National Institute of Allergy and Infectious Diseases (NIAID), the National Human Genome Research Institute (NHGRI), the National Institute of Child Health and Human Development (NICHD), the Juvenile Diabetes Research Foundation International (JDRF). In addition, the authors were supported by the European Foundation for the Study of Diabetes (Albert Renold Travel Grant- N Medina) and the Instituto de Salud Carlos III (PI11/02441).
Conflicts of interestThe authors are not aware of any conflicts of interest regarding the contents of this manuscript.
We would like to give our thanks to all the participating families who have made this study possible.
Francisco Javier Ampudia, Hospital Clínico de Valencia.
Jesús Argente, Hospital Niño Jesús, Madrid.
Luis Castaño, Hospital de Cruces, Baracaldo.
Raquel Corripio, Consorci Hospitalari Parc Taulí, Sabadell.
Mercè Fernández, Hospital Trueta, Girona.
Beatriz García Cuartero, Hospital Severo Ochoa, Leganés.
Concepción García Lacalle, Hospital Severo Ochoa, Leganés.
Pilar Gutiérrez, Hospital Universitario de Getafe.
Marta Hernández, Hospital Arnau de Vilanova, Lleida and Hospital Universitario de Canarias, Sta Cruz de Tenerife.
Alberto de Leiva, Hospital de la Santa Creu i Sant Pau, Barcelona.
Dídac Mauricio, Hospital Arnau de Vilanova, Lleida and Hospital de la Santa Creu i Sant Pau, Barcelona.
Francisco Javier Nóvoa, Hospital Universitario Insular de Gran Canaria.
Teresa Pedro, Hospital Clínico de Valencia, Mercedes Rigla, Hospital de la Santa Creu i Sant Pau, Barcelona.
María Jesús Rodríguez, Hospital Universitario Insular de Gran Canaria.
Mercedes Rodríguez, Hospital Miguel Servet, Zaragoza.
Óscar Rubio, Hospital Niño Jesús, Madrid.
Federico Vázquez, Hospital de Cruces, Baracaldo.
Ana María Wägner, Hospital Universitario Insular de Gran Canaria and Steno Diabetes Center, Denmark.