Only automated phenotypic methods are currently used in Colombian hospitals for identifying isolates of the Acinetobacter calcoaceticus–A. baumannii complex (ACB). The phenotypical similarities in these species mean that they cannot be differentiated by manual or automated methods, thereby leading to their identification as A. baumannii, or ACB complex in clinical settings. Our objective was to identify to the species level 60 isolates, from four hospitals, evaluate their antibiotic susceptibility, and detect resistance-related genes.
Methods16S–23S rRNA internal transcribed spacer (ITS) region and rpoB gene partial sequences were amplified. Resistance genes for cephalosporin, carbapenem and aminoglycoside were detected by PCR. Possible mutations in the quinolone resistance-determining region (QRDR) were evaluated. The association of ISAba-1 with blaOXA and blaADC genes was determined by PCR. Amplification products of ITS region, rpoB gene and some resistance genes were sequenced and compared using the BLAST tool.
Results16S–23S rRNA ITS region and partial rpoB gene sequence analysis allowed 51isolates to be identified as A. baumannii, 8 as A. nosocomialis, and 1 isolate as A. pitti. A. baumannii isolates were highly resistant to all antibiotics tested, while the others were susceptible to ciprofloxacin and ampicillin/sulbactam. Quinolone resistance, found only in A. baumannii, was associated with mutations in the QRDR region of gyrA and parC genes.
ConclusionThis is the first investigation in Colombia that has identified ACB complex species using molecular methods, and determined differences in antibiotic resistance and resistance genes among the species. It is of the highest importance to identify isolates to the species level for future resistance and epidemiology studies in our region.
Actualmente, los hospitales en Colombia utilizan únicamente métodos fenotípicos automatizados para la identificación de aislamientos del complejo Acinetobacter calcoaceticus – baumannii (ACB). La similitud entre estas especies no permite que se diferencien por métodos fenotípicos ya sean estos manuales o automatizados, llevando a que los aislamientos se identifiquen como A. baumannii o como pertenecientes al complejo ACB en las instituciones hospitalarias. Nuestro objetivo fue identificar a nivel de especie, 60 aislamientos de cuatro hospitales, identificados como del complejo ACB, evaluar su resistencia a antibióticos y detectar genes de resistencia.
MétodosPara la identificación de especies se amplificaron la región intergénica espaciadora de los genes 16S y 23S rRNA y la secuencia parcial del gen rpoB. Estos amplificados y algunos genes de resistencia se secuenciaron y se compararon utilizando la herramienta BLAST. Se detectaron por PCR genes de resistencia a cefalosporinas, carbapenemes y aminoglicósidos. Se evaluaron posibles mutaciones en la región determinante de resistencia a quinolonas (QRDR). Se determinó por PCR la asociación de ISAba-1con los genes blaOXA y blaADC.
ResultadosCon las secuencias de la región ITS 16S-23S rRNA y el gen rpoB, se identificaron 51 aislamientos como A. baumannii, 8 como A. nosocomialis y 1 como A. pittii. A. baumannii fue altamente resistente a todos los antibióticos ensayados. Las otras dos especies fueron susceptibles a ciprofloxacina y amplicilina/sulbactam. La resistencia a quinolonas se detectó únicamente en A. baumannii y se asoció con mutaciones en la región QRDR de los genes gyrA y parC.
ConclusionesEsta es la primera investigación en Colombia que identificó especies del complejo ACB usando métodos moleculares y determinó diferencias en la resistencia a antibióticos y en los genes de resistencia entre las especies. La identificación de los aislamientos a nivel de especie es de importancia para futuros estudios de resistencia y epidemiología en nuestra región.
Acinetobacter calcoaceticus–A. baumannii complex comprises four genomic species: A. calcoaceticus, A. baumannii, A. nosocomialis (formerly genomic species 13TU1) and A. pittii (formerly genomic species 31). Except for A. calcoaceticus, the other three species represent an important health threat as they are emerging as multidrug-resistant pathogens.2
As all these species are very close, genetically related, and difficult to identify by phenotypical methods using routine laboratory methods, it has been generally accepted to refer to them as a group (ACB complex),2 although differences among them regarding their antibiotic resistance, epidemiology and pathogenicity have been also well documented.3–5 Molecular methods as 16S–23S rRNA internal transcribed spacer (ITS) region analysis and rpoB gene fragment sequence analysis have been adopted for their identification.6
Regarding the antibiotic resistance, results from the SENTRY Antimicrobial Surveillance Program for Latin America and Brazil 1997–2001 indicate that resistant Gram-negative bacteria are much prevalent in Latin America than in North America and Europe. One of the main antimicrobial resistance problems is given by MDR (multidrug resistant) non-fermentative Gram-negative bacteria (Acinetobacter spp. and Pseudomonas aeruginosa).7 This problem is also referred by Casellas,8 indicating that species conforming ACB complex are resistant to a wide variety of antimicrobials as beta-lactams, fluoroquinolones, and aminoglycosides.
Previous studies carried out in Colombia focused on Acinetobacter spp. have shown high resistance to beta-lactams, aminoglycosides and quinolones, and an increasingly and concerning resistance to carbapenems.9,10
The purpose of this study was to establish by using molecular approaches, the species from representative ACB complex strains isolated from patients hospitalized in four Colombian hospitals, their antibiotic resistance patterns and the presence of genes associated with resistance to four different groups of antibiotics.
MethodsFrom an initial collection of 150 isolates obtained during 2004, 2005, 2007 and 2009 from hospitalized patients in 4 Colombian hospitals, we selected 60 isolates previously identified by automated systems (Vitek Biomérieux, Marcy-l’Etoile, France) as ACB complex isolates and with diverse antibiotic-resistance patterns. Isolates were tested for susceptibility to cefotaxime, ceftazidime, cefepime, imipenem, meropenem, aztreonam, ampicillin–sulbactam, piperacillin–tazobactam, amoxicillin–clavulanic acid, ciprofloxacin, amikacin, gentamicin and nalidixic acid by disk diffusion method, and using CLSI interpretive standards.11
All isolates were representatives of clones with less than 75% similarity as evaluated by repetitive extragenic palindromic PCR (REP-PCR). Some of them were representatives of clones with more than one isolates.
The 16S–23S rRNA internal transcribed spacer (ITS) region was amplified for each isolate and for the A. baumannii ATCC 19606 strain, according to the conditions established by Chang et al.6 The partial sequence of the rpoB gene was also amplified for all isolates that were not identified as A. baumannii and for some isolates identified by ITS as A. baumannii, according to conditions previously established.12
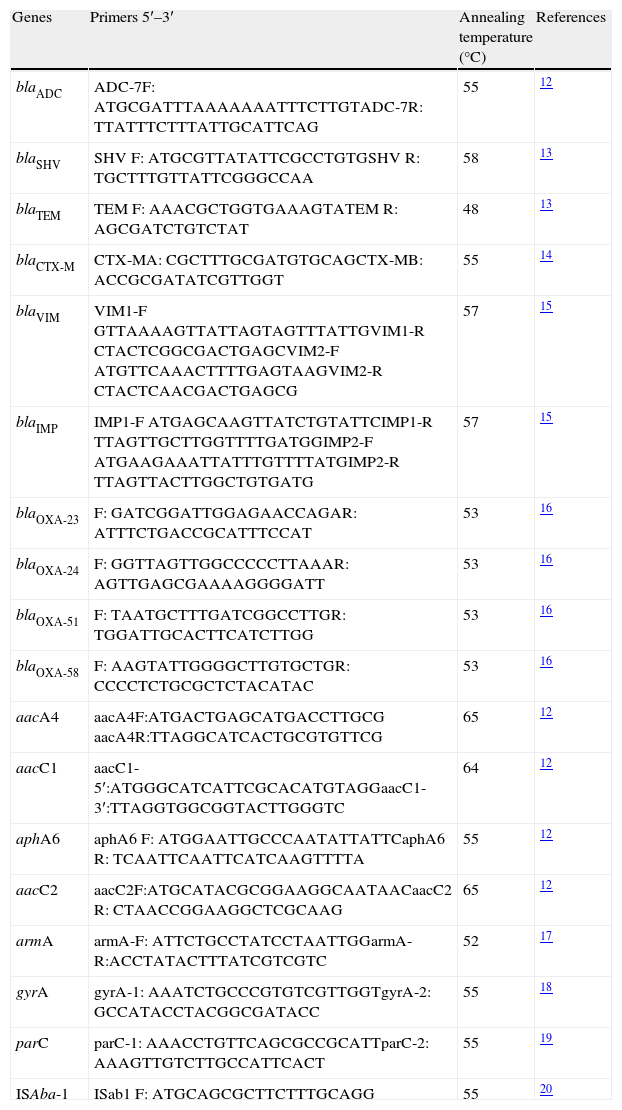
Genes codifying for resistance to cephalosporins, carbapenems and aminoglycosides were detected by PCR (Table 1). The strategy established by Vila et al. was used for detecting possible mutations in the quinolone resistance-determining region (QRDR).19,20 The association of ISAba-1 sequences with blaOXA and blaADC genes was also determined by PCR. Amplification was done with forward primer for insertion sequence and reverse primer for blaADC gene and blaOXA, respectively. Sequencing service was contracted with an external third party (Macrogen, Korea). Sequencing was done by single primer extension with an ABI Prism 3730xI-PE Applied Biosystems platform and resulting sequences were compared with sthose available on the Genbank.
Primers used for detection of resistance-related genes.
| Genes | Primers 5′–3′ | Annealing temperature (°C) | References |
| blaADC | ADC-7F: ATGCGATTTAAAAAAATTTCTTGTADC-7R: TTATTTCTTTATTGCATTCAG | 55 | 12 |
| blaSHV | SHV F: ATGCGTTATATTCGCCTGTGSHV R: TGCTTTGTTATTCGGGCCAA | 58 | 13 |
| blaTEM | TEM F: AAACGCTGGTGAAAGTATEM R: AGCGATCTGTCTAT | 48 | 13 |
| blaCTX-M | CTX-MA: CGCTTTGCGATGTGCAGCTX-MB: ACCGCGATATCGTTGGT | 55 | 14 |
| blaVIM | VIM1-F GTTAAAAGTTATTAGTAGTTTATTGVIM1-R CTACTCGGCGACTGAGCVIM2-F ATGTTCAAACTTTTGAGTAAGVIM2-R CTACTCAACGACTGAGCG | 57 | 15 |
| blaIMP | IMP1-F ATGAGCAAGTTATCTGTATTCIMP1-R TTAGTTGCTTGGTTTTGATGGIMP2-F ATGAAGAAATTATTTGTTTTATGIMP2-R TTAGTTACTTGGCTGTGATG | 57 | 15 |
| blaOXA-23 | F: GATCGGATTGGAGAACCAGAR: ATTTCTGACCGCATTTCCAT | 53 | 16 |
| blaOXA-24 | F: GGTTAGTTGGCCCCCTTAAAR: AGTTGAGCGAAAAGGGGATT | 53 | 16 |
| blaOXA-51 | F: TAATGCTTTGATCGGCCTTGR: TGGATTGCACTTCATCTTGG | 53 | 16 |
| blaOXA-58 | F: AAGTATTGGGGCTTGTGCTGR: CCCCTCTGCGCTCTACATAC | 53 | 16 |
| aacA4 | aacA4F:ATGACTGAGCATGACCTTGCG aacA4R:TTAGGCATCACTGCGTGTTCG | 65 | 12 |
| aacC1 | aacC1-5′:ATGGGCATCATTCGCACATGTAGGaacC1-3′:TTAGGTGGCGGTACTTGGGTC | 64 | 12 |
| aphA6 | aphA6 F: ATGGAATTGCCCAATATTATTCaphA6 R: TCAATTCAATTCATCAAGTTTTA | 55 | 12 |
| aacC2 | aacC2F:ATGCATACGCGGAAGGCAATAACaacC2 R: CTAACCGGAAGGCTCGCAAG | 65 | 12 |
| armA | armA-F: ATTCTGCCTATCCTAATTGGarmA-R:ACCTATACTTTATCGTCGTC | 52 | 17 |
| gyrA | gyrA-1: AAATCTGCCCGTGTCGTTGGTgyrA-2: GCCATACCTACGGCGATACC | 55 | 18 |
| parC | parC-1: AAACCTGTTCAGCGCCGCATTparC-2: AAAGTTGTCTTGCCATTCACT | 55 | 19 |
| ISAba-1 | ISab1 F: ATGCAGCGCTTCTTTGCAGG | 55 | 20 |
A. baumannii strain ATCC 19606 was used as a positive control in detection assays of the blaOXA-51 and blaADC genes, the ITS region, the rpoB gene fragment as well as a positive control for the restriction of the QRDR region of both gyrA and parC genes. Klebsiella pneumoniae strain 363 (from our laboratory) was used as a positive control for blaTEM, blaSHV and blaCTX-M genes.
PCR products from ITS region, rpoB gene, some resistance genes and gyrA and parC genes were sequenced in both chains in an external facility (Macrogen, Korea). Each sequence was compared with public databases using the BLAST tool (available at: http://blast.ncbi.nlm.nih.gov/Blast.cgi).
ResultsFifty-one out of the sixty isolates were identified as A. baumannii, eight as A. nosocomialis and one isolate was identified as A. pittii. ITS sequences revealed 99% to 100% similarity with those deposited in Genbank.
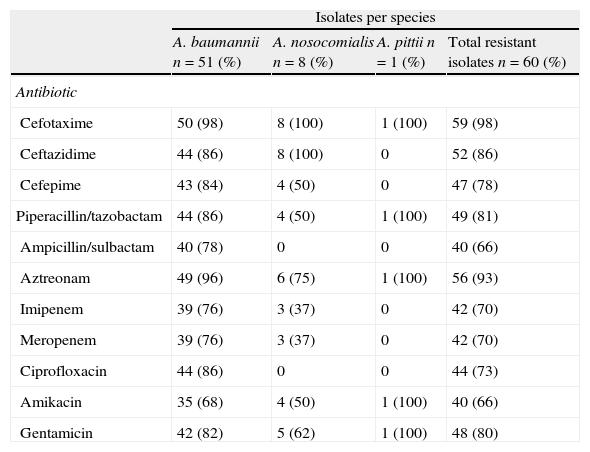
Isolates displayed different resistance patterns (Table 2). Antibiotics were grouped into six categories as follows: beta-lactams (cefotaxime, ceftazidime, cefepime, and aztreonam); carbapenems (imipenem, meropenem); beta-lactam/inhibitor combination (ampicillin–sulbactam, piperacillin–tazobactam, amoxicillin–clavulanic acid); quinolones (ciprofloxacin); aminoglycosides (amikacin, gentamicin) and nalidixic acid. According with definitions by Magiorakos et al.,21 46 isolates (all A. nosocomialis, the A. pittii and 37 A. baumannii) were classified as multidrug-resistant (MDR), 3 A. baumannii as extensively drug-resistant (XDR) and 10 A. baumannii as pandrug-resistant (PDR). Only one A. baumannii isolate was sensitive to all antibiotics tested except for nalidixic acid.
Antibiotic – resistance profile in Acinetobacter sp.
| Isolates per species | ||||
| A. baumannii n=51 (%) | A. nosocomialis n=8 (%) | A. pittii n=1 (%) | Total resistant isolates n=60 (%) | |
| Antibiotic | ||||
| Cefotaxime | 50 (98) | 8 (100) | 1 (100) | 59 (98) |
| Ceftazidime | 44 (86) | 8 (100) | 0 | 52 (86) |
| Cefepime | 43 (84) | 4 (50) | 0 | 47 (78) |
| Piperacillin/tazobactam | 44 (86) | 4 (50) | 1 (100) | 49 (81) |
| Ampicillin/sulbactam | 40 (78) | 0 | 0 | 40 (66) |
| Aztreonam | 49 (96) | 6 (75) | 1 (100) | 56 (93) |
| Imipenem | 39 (76) | 3 (37) | 0 | 42 (70) |
| Meropenem | 39 (76) | 3 (37) | 0 | 42 (70) |
| Ciprofloxacin | 44 (86) | 0 | 0 | 44 (73) |
| Amikacin | 35 (68) | 4 (50) | 1 (100) | 40 (66) |
| Gentamicin | 42 (82) | 5 (62) | 1 (100) | 48 (80) |
All A. nosocomialis were resistant to ceftazidime and aztreonam and susceptible to ampicillin/sulbactam and ciprofloxacin. A. pittii was resistant to five antibiotics and was sensitive to ceftazidime, cefepime, ampicillin/sulbactam, imipenem, meropenem and ciprofloxacin.
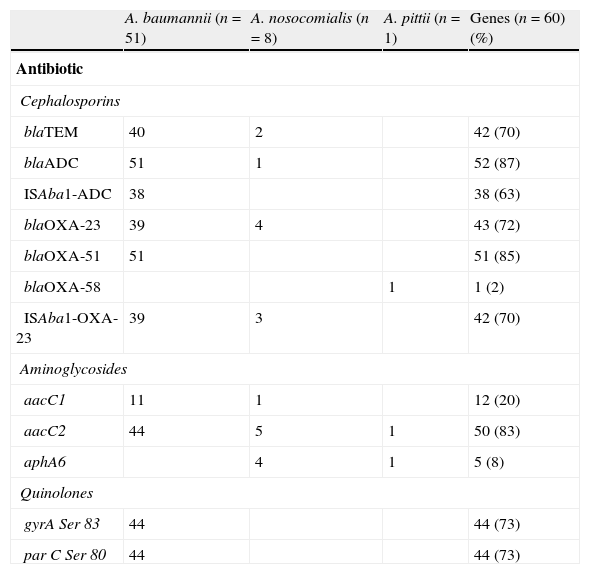
Regarding the betalactams resistance, genes detected were blaADC, blaTEM, and blaOXA. The blaADC gene was identified in all A. baumannii isolates (Table 3). Thirty-eight out of the 44 ceftazidime-resistant isolates had the ISAba-1 sequence upstream of this gene. The remaining (6/44) had the blaADC gene but no association with ISAba-1. Analysis of amplified blaADC sequences exhibited 100% of similarity with the blaADC-5 gene. Only one of the A. nosocomialis strains had the blaADC gene, but this was not associated with the ISAba-1 sequence. This gene was not detected in A. pittii. blaTEM genes were detected in 40 A. baumannii and in 2 A. nosocomialis isolates. The narrow spectrum TEM-1 enzyme-encoding gene was identified by sequence in some of these isolates. blaOXA genes from families 23, 51 and 58 were detected in the isolates studied; blaOXA-23 was detected in A. baumannii and A.nosocomialis and was found to be associated with the ISAba-1 sequence in all carbapenem-resistant isolates. blaOXA-23 was also detected in a carbapenem-sensitive A. nosocomialis strain, but the ISAba-1 sequence was not found upstream of the gene.
Resistance genes and QRDR mutations in Acinetobacter sp.
| A. baumannii (n=51) | A. nosocomialis (n=8) | A. pittii (n=1) | Genes (n=60) (%) | |
| Antibiotic | ||||
| Cephalosporins | ||||
| blaTEM | 40 | 2 | 42 (70) | |
| blaADC | 51 | 1 | 52 (87) | |
| ISAba1-ADC | 38 | 38 (63) | ||
| blaOXA-23 | 39 | 4 | 43 (72) | |
| blaOXA-51 | 51 | 51 (85) | ||
| blaOXA-58 | 1 | 1 (2) | ||
| ISAba1-OXA-23 | 39 | 3 | 42 (70) | |
| Aminoglycosides | ||||
| aacC1 | 11 | 1 | 12 (20) | |
| aacC2 | 44 | 5 | 1 | 50 (83) |
| aphA6 | 4 | 1 | 5 (8) | |
| Quinolones | ||||
| gyrA Ser 83 | 44 | 44 (73) | ||
| par C Ser 80 | 44 | 44 (73) | ||
The blaOXA-51 gene was only detected in A. baumannii and blaOXA-58 only in A. pittii. Those were not associated with ISAba-1.
Genes encoding aminoglycoside-modifying enzymes were detected: aacC2 (83.3%), aacC1 (20%) and aphA6 (8.3%). The aacC2 gene was found in members of the three species, most of which were resistant to gentamicin. The aphA6 gene was found exclusively in those amikacin-resistant species other than baumannii.
The 44 ciprofloxacin-resistant A. baumannii isolates presented mutations in codons 83 and 80 of gyrA and parC genes, respectively. The sequences of some of the amplified products which were not digested with Hinf1 had Ser 83 to Leu changes in gyrA and Ser 83 to Tyr changes in parC.
DiscussionSome authors consider that identification to the species level is necessary for monitoring resistance in each species and for establishing possible differences in clinical manifestations and outcomes for patients infected by Acinetobacter sp. belonging to the ACB complex.22
A. baumannii was the most frequently genomic species detected in the four hospitals studied, followed by A. nosocomialis. Only one isolate was identified as A. pittii. We found differences in resistance patterns according with the species in our study. A. baumannii isolates exhibited the three resistance categories (MDR, XDR and PDR), while some A. nosocomialis were susceptible to cefepime, beta-lactams/inhibitor combination, aminoglycosides and ciprofloxacin. Ciprofloxacin and ampicillin/sulbactam susceptibility observed in A. nosocomialis may give an option for the treatment of infections caused by this microorganism. This susceptibility in species other than baumannii coincided with that observed in other investigations.21,22 Presence of aphA6 gene exclusively in amikacin-resistant isolates of A. nosocomialis and A. pittii contrasted with what has been observed in other studies, where this gene has been found in a high percentage of A. baumannii.23 Resistance to third-generation cephalosporins was related to the ISAba-1-blaADC association in most of the isolates. This association has been especially documented in A. baumannii where such sequence provides a strong promoter for the overexpression of this gene.24,25
All the imipenem and meropenem resistant isolates had the ISAba-1-blaOXA 23 association. This constitutes the most disseminated mechanism of resistance to carbapenems in Acinetobacter sp. and some studies have shown that this association is enough to achieve resistance to this group of antibiotics. One A. nosocomialis that was sensitive to imipenem had the gene but it was not associated with the insertion sequence, thereby reaffirming the role of this IS in the overexpression of genes downstream of it.25
The blaOXA-51 gene was only identified in A. baumannii consistently with established by some authors who consider such gene to be species-specific in A. baumannii.22,26
The blaOXA-58 gene, was detected for the first time in Colombia only in the A. pittii identified in this study, although it has been previously detected in this species.27,28 The detection of OXA carbapenemases has been rare in species other than A. baumannii in previous studies.22 In this study the blaOXA genes were detected in the other two species.
All the ciprofloxacin-resistant A. baumannii strains presented mutations in QRDR codons 83 and 80 in gyrA and parC genes and this is the main mechanism associated with resistance to quinolones as previously reported in other studies.19,20
High resistance to aminoglycosides was observed in the three species and the aacC2 gene was associated with resistance to gentamicin. The aacC1 gene was detected only in resistant isolates and concomitantly with aacC2 genes. The aphA6 gene, which confers resistance to amikacin and gentamicin, was only found in A. nosocomialis and A. pittii that were resistant to both gentamicin and amikacin. Resistance to amikacin in A. baumannii however, could not be explained with this gene evaluation. Other genes encoding aminoglycoside-modifying enzymes, as well as other non-enzymatic mechanisms, could be responsible for resistance to amikacin in A. baumannii. The absence of aphA6 gene in our A. baumannii strains contrasted with those from European clones I and II where this gene is found together with other aminoglycoside-resistant ones.23
This is the first report in Colombia that use molecular methods for differentiating species from the ACB complex and establish the presence of A.nosocomialis and A. pittii in hospitals in Colombia. The differences found in species susceptibility and molecular basis of that resistance, lead us to recommend that molecular techniques should be implemented in studies of surveillance of antibiotic resistance in members of the ACB complex.
FundingFinancial resources for this research came from Departamento Administrativo de Ciencia, Tecnología e Innovación – Colciencias (contrato 110145221066) and Universidad Nacional de Colombia, Dirección de Investigación– sede Bogotá (DIB).
Conflicts of interestAuthors state that there does not exist any personal or financial relationship that may have caused a conflict of interest regarding this manuscript.
Authors wish to thank Departamento Administrativo de Ciencia, Tecnología e Innovación – Colciencias (contrato 110145221066) and Universidad Nacional de Colombia, Dirección de Investigación– sede Bogotá (DIB), for the finantial support to this research.
Thanks also to the microbiology teams of the participating hospitals for supplying the bacterial isolates and for their valuable contributions to this work.