The epidemiology of Burkholderia cepacia complex (Bcc) in cystic fibrosis (CF) is not widely known.
MethodsAll CF patients with Bcc between 2002 and 2011 were reviewed, and a molecular analysis of isolates was performed.
ResultsThe prevalence of Bcc infection was 7.2% (18/250). Molecular analysis of 16 Bcc isolates showed 5 species (7 B. contaminans, 6 B. cepacia, 1 B. cenocepacia, 1 B. multivorans, and 1 B. stabilis) and 13 sequence types. There were no cases of cross-transmission.
ConclusionA high diversity of Bcc species was found in infected CF patients.
La epidemiología de Burkholderia cepacia complex (Bcc) en fibrosis quística (FQ) no es bien conocida.
MétodosSe revisaron todos los pacientes con Bcc entre 2002-2011. Se realizó análisis molecular de los aislamientos.
ResultadosLa prevalencia fue de 7.2% (18/250). El análisis molecular de 16 aislamientos representativos mostró 5 especies (7 B. contaminans, 6 B. cepacia, 1 B. cenocepacia, 1 B. multivorans, and 1 B. stabilis), y 13 tipos de secuencia. No hubo casos de transmisión cruzada.
ConclusionesSe encontró una alta diversidad de especies de Bcc causantes de infecciones en FQ.
Burkholderia cepacia complex (Bcc) is a closely related group of 17 bacterial species with similar phenotype but different genotype.1 Colonization with Bcc is associated with a constant and accelerated decline in lung function and increased risk of death in cystic fibrosis (CF) patients. Particular species of the Bcc such as Burkholderia cenocepacia and Burkholderia multivorans have been associated with an increased transmission risk and virulence, and with a worse outcome following lung transplantation.2
In Spain, the prevalence of chronic Bcc infection in CF patients in 2009 was 3.5% (data collected from only 30% of the total CF population in Spain).3 The distribution of Bcc species for this population is not well established, except for a recent report stating that B. cenocepacia was the most prevalent genomovar found in patients with CF (19.1%).4 An increase in the number of CF Bcc cases in our center was identified during 2009–2010, for which, a decision was taken to investigate the clinical characteristics of CF patients with Bcc, the distribution of Bcc species, and the possibility of cross-transmission among the patients treated in our CF unit.
MethodsA retrospective study was performed on all CF patients colonized/infected with Bcc isolates from January 2002 to December 2011 treated in the CF unit of the Hospital Universitario 12 de Octubre (Madrid). Clinical records were reviewed in order to collect demographic, clinical, and microbiology data. Stringent infection control measures were implemented to reduce cross-transmission, and included hand washing, use of gloves and masks, disposable mouthpieces and antimicrobial filters when a spirometry was performed.
Sputum samples were inoculated onto Columbia 5% blood, MacConkey, mannitol-salt, chocolate, and B. cepacia (BCSA) agar medium. The identification and the susceptibility to different antimicrobials were carried out using the microdilution method (Wider® System, Soria-Melguizo, Spain) and ¿-test strips (AB, Biodisk, Solna, Sweden) for ceftazidime (CAZ), meropenem (MER), levofloxacin (LVX), minocycline (MC), chloramphenicol (CLO) and trimethoprim-sulfamethoxazole (SXT). The minimum inhibitory concentrations (MICs) were interpreted according to the Clinical Laboratory Standards Institute.5
Bcc strains were genotyped by pulsed-field gel electrophoresis (PFGE) with the restriction enzyme XbaI.6 PCR amplification of recA7 gene was used to identify at species level. Multilocus sequence typing8 (MLST) was performed, besides to establish clonality, to confirm the identification. Sequencing reactions were run on an ABI Prism 3100 Genetic Analyzer, obtaining the taxonomic status by sequence analysis using GenBank and the B. cepacia MLST database (http://pubmlst.org/bcc/). Novel alleles and STs were submitted. Univariate analysis was performed by using the t test for continuous variables and the χ2 or Fisher exact tests for categorical variables.
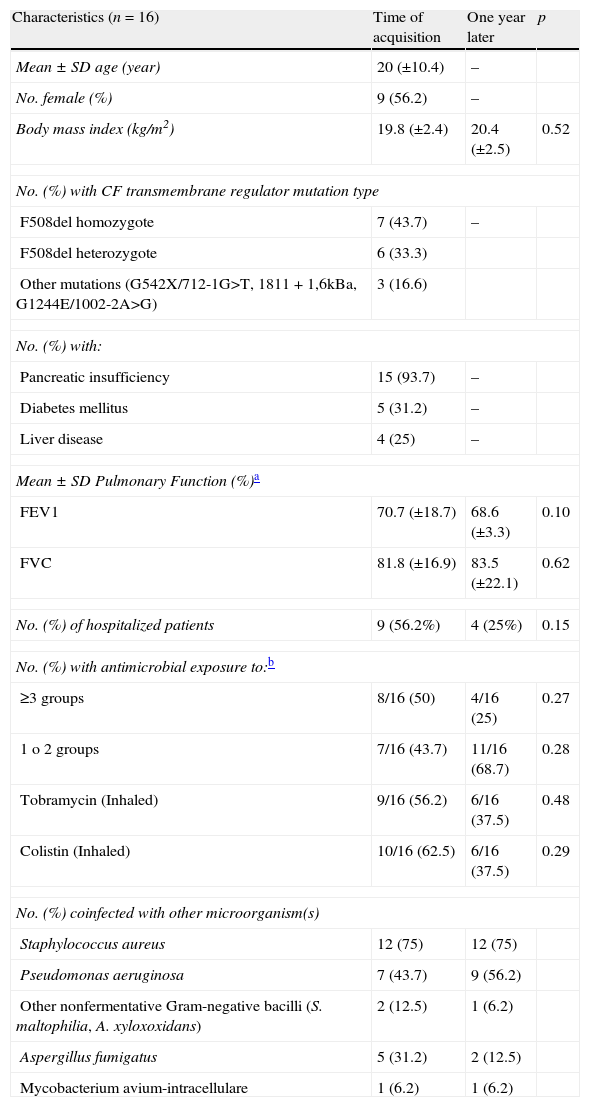
ResultsOver the 10-year period of study, 70 Bcc isolates putatively identified by phenotypic methods from 18 CF patients were recovered from a total of 250 patients treated in our CF unit. The prevalence of Bcc infection was 7.2%. Clinical isolates from two patients were not available, and therefore removed from the study. Nine patients were considered as chronically colonized (at least three positive cultures obtained in one year) and seven as sporadically colonized (less than three positive cultures). Ten (62.5%) cases were detected in 2009–2010, and 12 patients (75%) were older than 16 years. Clinical parameters for the 16 Bcc infected patients did not demonstrate any significant trends with body mass index, lung function, hospitalization and antimicrobial usage unchanged after one year of infection (Table 1). We also compared these clinical parameters between chronic and sporadic colonizers, and we could not establish a relationship between Bcc infection and a worse outcome in the lung function, probably because we only analyzed the outcome one year after the first isolation, and this limited the power of this analysis. It should be noted that one patient with chronic ST208 B. cenocepacia infection, had lung transplantation. Following this, the patient had two episodes of cepacia syndrome, and finally, died after 8 months.
Baseline characteristics of patients with Burkholderia cepacia complex infection at time of acquisition and one year after the first isolation.
| Characteristics (n=16) | Time of acquisition | One year later | p |
| Mean±SD age (year) | 20 (±10.4) | – | |
| No. female (%) | 9 (56.2) | – | |
| Body mass index (kg/m2) | 19.8 (±2.4) | 20.4 (±2.5) | 0.52 |
| No. (%) with CF transmembrane regulator mutation type | |||
| F508del homozygote | 7 (43.7) | – | |
| F508del heterozygote | 6 (33.3) | ||
| Other mutations (G542X/712-1G>T, 1811+1,6kBa, G1244E/1002-2A>G) | 3 (16.6) | ||
| No. (%) with: | |||
| Pancreatic insufficiency | 15 (93.7) | – | |
| Diabetes mellitus | 5 (31.2) | – | |
| Liver disease | 4 (25) | – | |
| Mean±SD Pulmonary Function (%)a | |||
| FEV1 | 70.7 (±18.7) | 68.6 (±3.3) | 0.10 |
| FVC | 81.8 (±16.9) | 83.5 (±22.1) | 0.62 |
| No. (%) of hospitalized patients | 9 (56.2%) | 4 (25%) | 0.15 |
| No. (%) with antimicrobial exposure to:b | |||
| ≥3 groups | 8/16 (50) | 4/16 (25) | 0.27 |
| 1 o 2 groups | 7/16 (43.7) | 11/16 (68.7) | 0.28 |
| Tobramycin (Inhaled) | 9/16 (56.2) | 6/16 (37.5) | 0.48 |
| Colistin (Inhaled) | 10/16 (62.5) | 6/16 (37.5) | 0.29 |
| No. (%) coinfected with other microorganism(s) | |||
| Staphylococcus aureus | 12 (75) | 12 (75) | |
| Pseudomonas aeruginosa | 7 (43.7) | 9 (56.2) | |
| Other nonfermentative Gram-negative bacilli (S. maltophilia, A. xyloxoxidans) | 2 (12.5) | 1 (6.2) | |
| Aspergillus fumigatus | 5 (31.2) | 2 (12.5) | |
| Mycobacterium avium-intracellulare | 1 (6.2) | 1 (6.2) | |
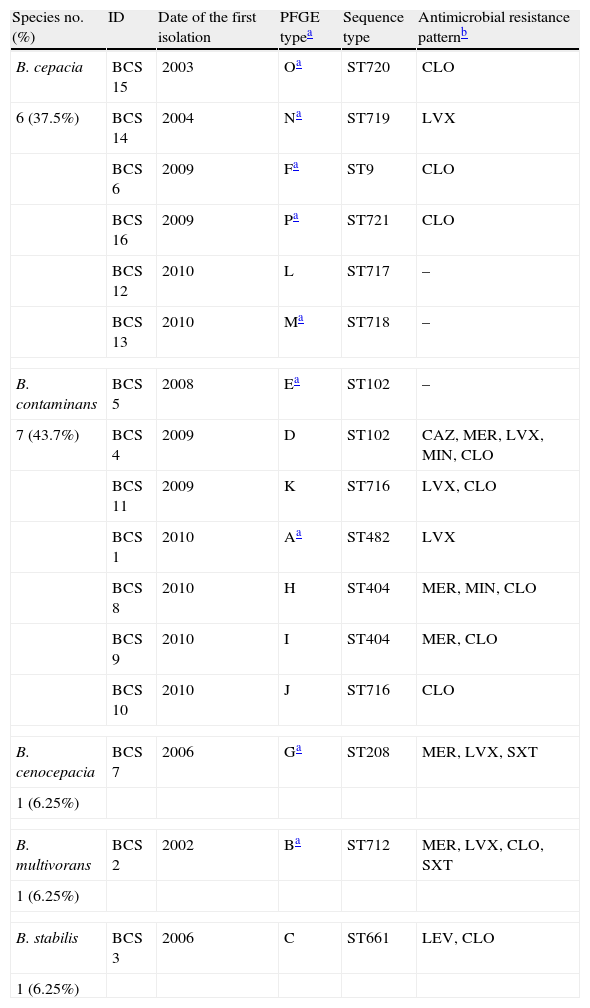
A total of 41 Bcc isolates phenotypically identified were available for molecular studies: 34 isolates from 9 chronically colonized patients and 7 from 7 sporadically colonized patients. recA PCR-sequencing and MLST were performed in 16 Bcc isolates (one per patient) and showed 5 different species and 13 different sequence types (STs): 6 B. cepacia (ST9, ST717, ST718, ST719, ST720, ST721), 7 B. contaminans (ST102 [2], ST404 [2], ST482, ST716 [2]), 1 B. cenocepacia (ST208), 1 B. multivorans (ST712), and 1 B. stabilis (ST661) (Table 2). Six were novel STs (ST716, ST717, ST718, ST719, ST720, and ST721).
Distribution of Burkholderia cepacia complex species according to the PFGE patterns, sequence types and antimicrobial resistance patterns.
| Species no. (%) | ID | Date of the first isolation | PFGE typea | Sequence type | Antimicrobial resistance patternb |
| B. cepacia | BCS 15 | 2003 | Oa | ST720 | CLO |
| 6 (37.5%) | BCS 14 | 2004 | Na | ST719 | LVX |
| BCS 6 | 2009 | Fa | ST9 | CLO | |
| BCS 16 | 2009 | Pa | ST721 | CLO | |
| BCS 12 | 2010 | L | ST717 | – | |
| BCS 13 | 2010 | Ma | ST718 | – | |
| B. contaminans | BCS 5 | 2008 | Ea | ST102 | – |
| 7 (43.7%) | BCS 4 | 2009 | D | ST102 | CAZ, MER, LVX, MIN, CLO |
| BCS 11 | 2009 | K | ST716 | LVX, CLO | |
| BCS 1 | 2010 | Aa | ST482 | LVX | |
| BCS 8 | 2010 | H | ST404 | MER, MIN, CLO | |
| BCS 9 | 2010 | I | ST404 | MER, CLO | |
| BCS 10 | 2010 | J | ST716 | CLO | |
| B. cenocepacia | BCS 7 | 2006 | Ga | ST208 | MER, LVX, SXT |
| 1 (6.25%) | |||||
| B. multivorans | BCS 2 | 2002 | Ba | ST712 | MER, LVX, CLO, SXT |
| 1 (6.25%) | |||||
| B. stabilis | BCS 3 | 2006 | C | ST661 | LEV, CLO |
| 1 (6.25%) | |||||
PFGE analysis of the 41 Bcc isolates showed 16 different pulsotypes. Furthermore, those belonging to the same species (B. cepacia and B. contaminans) had different DNA patterns (Table 2). Thirty-four sequential isolates recovered from 9 patients (range 10 months–4 years) were analyzed by PFGE, and all patients harbored the same strain over the time.
The percentages of resistance to antimicrobials were: CAZ (6.2%), MER (31.2%), LVX (37.5%), MC (12.5%), CLO (72.5%) and SXT (12.5%). The MICs were: CAZ (range 1–256μg/ml, MIC90%: 8μg/ml, MIC50%: 4μg/ml), MER (range 0.38–32μg/ml, MIC90%: >32μg/ml, MIC50%: 1.5μg/ml), LVX (range 0.38–32μg/ml, MIC90%: 6μg/ml, MIC50%: 1.5μg/ml), MC (range 0.19–32μg/ml, MIC90%: 6μg/ml, MIC50%: 3μg/ml), CLO (range 0.5–32μg/ml, MIC90%: 32μg/ml, MIC50%: 12μg/ml) and SXT (range 0.047–32μg/ml, MIC90%: 6μg/ml, MIC50%: 0.38μg/ml).
DiscussionThis study detected a prevalence of 7.2% of Bcc infection in CF patients. Other European countries and United States have reported rates ranged between 0 and 11.5%.3,9 The most prevalent species were B. contaminans (43.7%) and B. cepacia (37.5%). B. cenocepacia and B. multivorans represented 12.5%. However, other European countries, Australia, New Zealand, Canada, and United States, reported rates for B. cepacia between 0 and 11.2%, for B. cenocepacia between 45.1 and 91.8% and for B. multivorans between 0 and 51.6%.10–12 A high clonal diversity was also found in the two more frequent species, B. contaminans and B. cepacia. B. contaminans belonging to ST102 has been involved in a widespread outbreak in the United States and Brazil in non-cystic fibrosis patients.13 In Spain, ST102 has been reported as cause of an outbreak of subclinical mastitis in dairy sheep.1,13,14 The B. cenocepacia isolate of this study did not belong to either the epidemic ET12 strain (ST28) or the Czech strain (ST32), which have been associated with serious outbreaks among patients in Canada and Europe.10 Portugal seems to be the scenario where B. cepacia have been seen to be dominant in CF patients.15 Since the environment will be source of Bcc infection in the absence of strain transmission, then we could speculate that environmental sources of B. cepacia may be more common in Portugal and Spain, and hence lead to it being dominant in CF in these two close countries. Differences in the prevalence of Bcc species, at the regional and local level, may result from a combination of several factors, as genetic diversity among Bcc species living in the natural environment, predominance of epidemic strains (belonging mostly to B. cenocepacia), antimicrobial pressure, and adherence to infection control measures.10
Cases of cross-transmission among CF patients were not found although 62.5% of cases were detected in the last two years of study. Although the increase in 2009–2010 was remarkable, there were not changes in handling, processing or identification procedures in our laboratory that explained the increase in the number of Bcc cases. Probably, it could be related in part to having started to use inhaled colistin in our CF unit from 2005 (data not shown). We also investigated if the Bcc strains could persist in the same patient causing infection. In 9 patients, chronic infection involved a single strain. Bernhardt et al. found that replacement of the initial infecting strain occurred in 6.9% of patients infected with Bcc strains.16
Although our study refers only to a single CF unit, it illustrates the high diversity of Bcc species infecting CF patients. Given the great divergence in the different countries, it is important to know the local epidemiology due to the impact of this information on the clinical management of patients and on the implementation of appropriate infection control policies.
Conflict of interestThe authors declare no conflict of interest.
We thank Dr. Joaquín R. Otero for reviewing the manuscript.
This study was supported by the Spanish Network for the Research in Infectious Diseases (RD06/0008) from the Instituto de Salud Carlos III and Fundación Mutua Madrileña (FMM 2011/0064).