Onychomycosis is a frequent and underdiagnosed condition. Approximately 90% of toenail onychomycosis infections are caused by dermatophytes, but classical diagnosis based on culture and microscopy observation is slow and has low sensitivity. Both limitations can be solved incorporating molecular techniques to routine diagnosis of onychomycosis.
ObjectiveProspective evaluation of the utility of incorporating in the clinical laboratory workflow a commercial real time PCR (qPCR) for dermatophytes detection in nails after potassium hydroxide direct observation screening.
Materials and methods152 nail samples were included (34 KOH negative and 118 KOH positive) and processed by culture and qPCR.
ResultsIn the negative KOH group, only one dermatophyte grew in culture and three were detected by qPCR. In the group of positive KOH, 57 dermatophytes grew in culture and 81 were detected by qPCR. In this group, 25% of diagnosed dermatophytes were detected only by qPCR. The sensitivity of qPCR compared to culture is 92.8% and time of response decreases from days to hours.
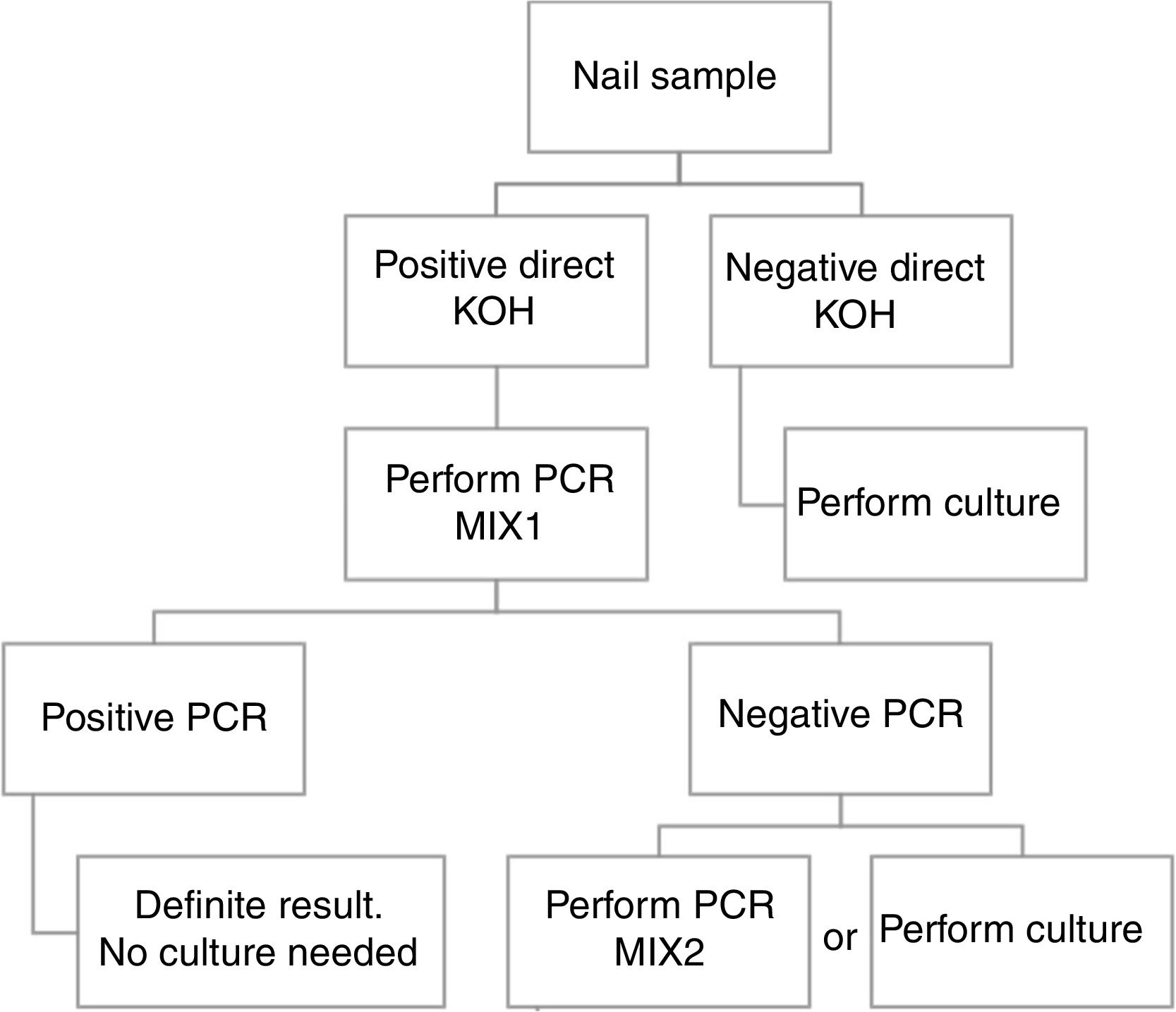
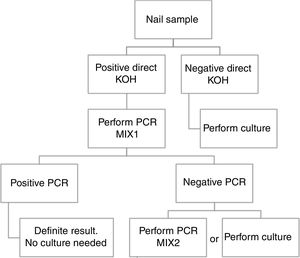
ConclusionBased in our results, we propose a workflow algorithm for a clinical laboratory that eliminates culture for qPCR positive samples.
La onicomicosis es una patología frecuentemente infradiagnosticada. Aproximadamente el 90% de las infecciones en las uñas del pie están causadas por dermatofitos, pero el diagnóstico microbiológico clásico basado en cultivo y microscopia es lento y tiene una baja sensibilidad. Ambas limitaciones pueden resolverse incorporando técnicas moleculares al diagnóstico de la onicomicosis.
ObjetivoEvaluación prospectiva de la utilidad de incorporar en un laboratorio clínico una PCR a tiempo real (qPCR) comercial para detección de dermatofitos en uñas tras cribado por examen directo con hidróxido de potasio (KOH).
Materiales y métodosSe incluyeron 152 muestras de uñas (34 KOH negativas y 118 KOH positivas) y se procesaron mediante cultivo y qPCR.
ResultadosEn el grupo KOH negativo, solo un dermatofito creció en cultivo y 3 se detectaron mediante qPCR. En el grupo KOH positivo, 57 dermatofitos crecieron en cultivo y 81 se detectaron por qPCR. En este grupo, el 25% de los dermatofitos diagnosticados se detectaron únicamente mediante qPCR. La sensibilidad de la qPCR comparada con el cultivo es del 92,8% y el tiempo de respuesta disminuye de días a horas.
ConclusiónEn base a nuestros resultados, proponemos un algoritmo de flujo de trabajo para los laboratorios de microbiología clínica, que elimina el cultivo para aquellas muestras con qPCR positiva.
Onychomycosis is a nail infection that can be caused by dermatophytes, non-dermatophyte moulds, and yeasts. Approximately 90% of toenail onychomycosis infections are caused by dermatophytes (e.g., Trichophyton, Microsporum, and Epidermophyton) with Trichophyton rubrum and T. interdigitale the most common causal strains.1
Dermatophytes are a group of keratinolytic filamentous fungi that invade keratinized tissues causing mostly superficial infections involving the skin, hair and nails. They are amongst the most common causes of skin disease in the world, and the real prevalence is probably underestimated.2
Onychomicosis is not a serious condition, but it affects patients comfort in various aspects: aesthetic, frequent medical consultation, long evolution, etc. and makes the patient feel stigmatized for its popular association with poor hygiene and contagious risk.3
Diagnosis of onychomycosis is usually made by combination of culture and direct examination with KOH, but they have several limitations. Both of them have low sensitivity and culture is slow and requires the microorganism to be alive.4 Moreover, the isolation of various non-dermatophyte filamentous fungi (NDF) such as Fusarium spp., Aspergillus spp. and Alternaria spp. is frequent but their role in nail infection can only be suspected with some certainty in repeated isolations,5 which increases the time until diagnosis and may cover up a dermatophyte infection.
This scenery requires efficient dermatophyte diagnosis techniques. Although culture is considered the gold standard for onychomycosis diagnosis, it is known that it is an imperfect reference, due to its comparably low sensitivity.6 Molecular techniques (e.g., qPCR) allow faster and more sensitive diagnosis, with the advantage of detection of non-viable microorganisms.4
First molecular methods were in-house assays that could be difficult to implement in routine laboratories because they require skilled technicians,7 usually detect only one pathogen per reaction and are sometimes difficult to reproduce.2 This challenge has recently been overcome with the introduction of several commercial kits that have been validated for the detection of dermatophytes in nails. One of them is DermaGenius® 2.0 (PathoNostics, The Netherlands),7 a multiplex real time PCR (qPCR) that can be performed in any Molecular Microbiology laboratory. The whole procedure takes around 3h and interpretation must be made for any person who is familiar with this techniques.
The aim of this study is to evaluate the utility of performing a commercial dermatophyte real time PCR in nail samples to increase the diagnosis of dermatophyte-caused onychomycosis and decrease the time of response until a positive result is given and also to design an algorithm to implement this qPCR in a clinical laboratory.
Materials and methodsClinical samples received in Microbiology Clinical laboratory for onychomycosis study between March and June 2019 were included in the study.
All nail samples were examined microscopically. A portion of the sample was covered with a solution of potassium hydroxide (KOH) 10% and incubated for at least 3h before examination. A KOH was considered positive if structures compatible with yeast or filamentous fungi were observed, and qPCR was performed. For every 3 positive KOH samples, one negative KOH sample was included in the qPCR study.
All samples were cultured in Sabouraud Gentamicine Chloramphenicol 2 (bioMérieux) and Dermatophyte solid agar (bioMérieux) (30°C, aerobic conditions for 21 days) and examined once a week. If fungal growth was detected, identification was made using MALDI-TOF MS (VITEK MS version 3.2, bioMérieux,) using VITEK® MS MOULD extraction kit.
Direct nail samples qPCR (DermaGenius® 2.0, PathoNostics, The Netherlands) was performed using a CFX96 device (BioRad) following the manufacturer's instructions, both for nucleic acids extraction and amplification steps. Both supplied mixes were used. MIX 1 detects C. albicans, T. tonsurans, T. mentagrophytes, T. rubrum/soudanensi, T. interdigitale and T. violaceum and MIX 2 detects T. benhamiae, T. verrocusum, Microsporum audouinii, M. canis and Epidermophyton floccosum.
Results152 nail samples were included in the study. Of them, 34 had negative direct KOH and 118 had positive direct KOH.
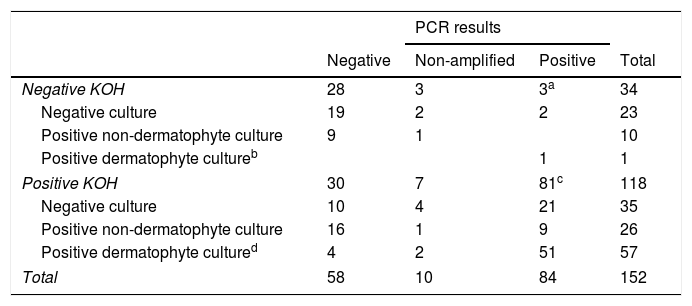
In the group of negative KOH samples, 11/34 had a positive culture: 1 T. rubrum and 10 moulds and yeasts others than C. albicans (4 Fusarium spp., 2 C. parapsilosis, 1 Alternaria spp. and 3 others). 3/34 (8%) had a positive qPCR result (3 T. rubrum/soudanensi), 28/34 had a negative qPCR result and 3/34 were non-amplifiable (Table 1).
PCR results according to KOH and culture result.
| PCR results | ||||
|---|---|---|---|---|
| Negative | Non-amplified | Positive | Total | |
| Negative KOH | 28 | 3 | 3a | 34 |
| Negative culture | 19 | 2 | 2 | 23 |
| Positive non-dermatophyte culture | 9 | 1 | 10 | |
| Positive dermatophyte cultureb | 1 | 1 | ||
| Positive KOH | 30 | 7 | 81c | 118 |
| Negative culture | 10 | 4 | 21 | 35 |
| Positive non-dermatophyte culture | 16 | 1 | 9 | 26 |
| Positive dermatophyte cultured | 4 | 2 | 51 | 57 |
| Total | 58 | 10 | 84 | 152 |
a,bTrichophyton rubrum/soudanensi. c 71 Trichophyton rubrum/soudanense, 6 Trichophyton interdigitale, 1 Trichophyton rubrum/soudanense and Epidermophyton flocossum co-detection. d 47 Trichophyton rubrum, 5 Trichophyton interdigitale, 3 Trichophyton violaceum, 2 Candida albicans.
In the group of positive KOH samples, 83/118 had a positive culture: 47 T. rubrum, 5 T. interdigitale, 3 T. violaceum, 2 C. albicans, 26 moulds and yeasts other than C. albicans (8 Aspergillus spp., 3 Alternaria spp., 3 Fusarium spp., 2 C. parapsilosis, 2 C. guillermondii, 2 C. famata and 6 others). 81/118 had positive qPCR result (3 C. albicans, 6 T. interdigitale, 71 T. rubrum/soudanensi and 1 co-detection of T. rubrum/soudanensi and E. flocossum), 30/118 had a negative qPCR result and 7/118 were non-amplifiable (Table 1).
Only one dermatophyte included in MIX 2 (E. flocossum) was detected in a single sample, in which T. rubrum/soudanense (MIX 1) was co-detected. In the culture of this sample, only T. rubrum was isolated.
In four cases of positive T. rubrum/soudanense qPCR, MALDI-TOF MS identification was T.violaceum and another colony was identified as 50% T. rubrum/50% T.violaceum.
In four cases of positive T. rubrum/soudanense qPCR, a filamentous fungi was isolated in culture, but MALDI-TOF was not able no identify them and microscopic examination with lactofenol-blue was not conclusive since the colonies did not sporulate.
21/35 KOH positive samples with negative culture and 9/26 KOH positive samples with positive culture for moulds or yeast other than C. albicans had a positive dermatophyte result by qPCR.
Overall, in the group of negative KOH, qPCR allowed to give an additional 5% (2/34) of positive results; however, in the group of positive KOH, 25% (30/118) of dermatophyte positive results were given thanks to qPCR.
The mean time of response for positive culture was 13.5±5.01 days while qPCR results were available in approximately 3h.
Using conventional culture as the gold standard, DermaGenius®2.0 qPCR demonstrated 92.8% sensitivity, 62.7% specificity, 61.9% VPP and 93.1% VPN.
DiscussionOnychomycosis is a frequent nail infection caused by filamentous fungi, primarily dermatophytes. Fungal infections diagnosis based in traditional culture is slow. In this work, we evaluate the introduction of a real time PCR for dermatophyte detection in nail samples.
Overall, qPCR allowed the etiologic diagnosis of dermatophyte-caused onychomycosis in 55.2% of cases (84/152), while culture only accomplished 38.1% (58/152) of diagnosis.
Hayette et al. performed a similar study also using DermaGenius® 2.0 qPCR. They evaluated this commercial qPCR in 138 nail samples in comparison to histology and culture results. When comparing their study to ours, the rate of dermatophytes detected both for culture and qPCR is almost the same (37.6% vs 34.2%) and also is the rate of false negative results (5% vs 6.7%). The recovery rate of dermatophytes by qPCR that did not grow in culture was similar in both studies, (39.7% vs 34%).
Spiliopoulou4 and Kondori8 described also important increases in the recovery of dermatophytes using other commercial multiplex PCR, with PCR always performing better than culture.
It is known that culture has a low sensitivity. Considering culture as the gold standard, all positive qPCR with negative culture or non-dermatophyte isolation must be considered false positive. For this reason, qPCR specificity and VPP yielded biased low values. However, if the gold standard is changed to culture combined with qPCR, DermaGenius® 2.0 qPCR demonstrated 93.3% of sensitivity, while culture exhibited only 64.4%. Others authors have already described similar results.3,4
In the present study, we detected 4 PCR false negative results. It is known that this may be due to the presence of PCR inhibitors in the sample,7 but in our case the internal control was correctly amplified, what means that there was no inhibition of the PCR.
It has been described that the irregular distribution of fungal elements within the sample might be a cause of false negative results4,6 and this seems the most probable explanation in our case.
On the other hand, we obtained 10 non-amplifiable results that yielded the same result when repeated with a new piece of sample. It seems that some PCR inhibitor must be present within the sample. In this situation, PCR repetition should be avoided and we recommend to perform conventional culture.
Note that we decided to evaluate the qPCR using both MIX 1 and MIX 2. We only detected one microorganism corresponding to MIX 2 (E. flocossum) and it was in co-detection with another one from MIX 1 (T. rubrum). Although manufacturer's instructions recommend using only MIX 1 for nail samples, and in our study none of the diagnosis of onychomycosis would have been missed, we think that using MIX2 for those cases of positive KOH and negative MIX1 qPCR would be of interest.
Although MALDI-TOF is a good tool for identification of dermatophytes,9 misidentification of T. soudanense (the African variant of T. rubrum) as T. violaceum when using Vitek MS V2.0.0 database has been described.9,10 In our study, we detected this discrepancy in four samples with a culture identified as T. violaceum by MALDI-TOF that were correctly identified as T.rubrum/soudanense thanks to qPCR.
Non-dermatophyte filamentous fungi are usually isolated in onychomycosis cultures. This kind of microorganisms grow quickly and can inhibit the growing of dermatophytes, but its clinical value is frequently uncertain. When incorporating qPCR, dermatophytes were detected in 25% of these samples that would have been otherwise informed as uncertain clinical value and with a recommendation of repeating the culture.
In addition to the increase of dermatophyte detection, it is of great importance the decrease of time of response. We have a mean time of response of 13 days for conventional culture and it decreases to hours when qPCR is used. The saving of agar plates, residue generation and time of examination and identification, must be taken into account when evaluating the implementation of a molecular technique.
Based in our results, we propose an algorithm to PCR performance (Fig. 1). First a direct KOH screening for all the nail samples. For those with a positive result, DermaGenius® 2.0 qPCR may be performed using MIX 1. KOH screening is of high importance in order to increase the profitability of the PCR. Verrier et al.5 described a high percentage of negative PCR results attributed to the lack of previous microscopic examination to select positive samples.
Routine study of dermatophyte antifungal susceptibility is not recommended. For this reason, culture of positive qPCR samples could be not necessary.
The implementation of molecular techniques with a validated algorithm in a particular scenario, could help avoiding false negative cultures or uncertain clinical significance results, which usually ends in incorrect or unnecessary treatments and a repeated visits to the primary health office.
Conflict of interestThe authors report no conflicts of interest.