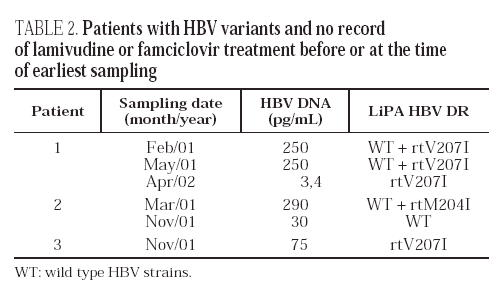Introduction
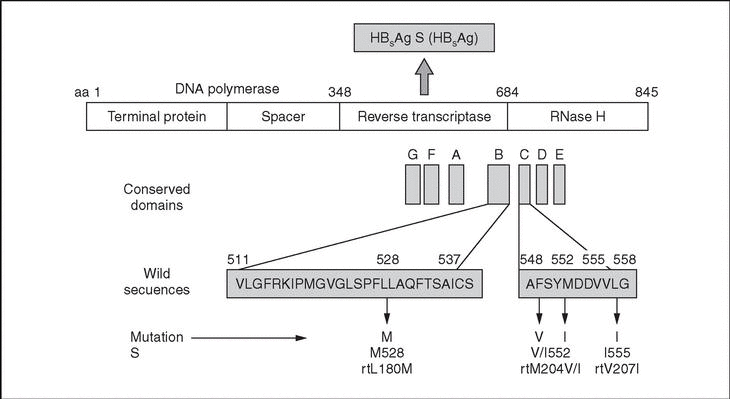
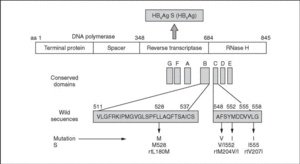
Lamivudine is an antiviral drug approved for the treatment of chronic hepatitis B. Lamivudine therapy is well tolerated and is effective in reducing hepatitis B virus (HBV) replication and inducing normalization of serum ALT levels. Nevertheless, selection of mutant viral strains resistant to the drug is common, occurring at a rate of around 20% per year of treatment among immunocompetent patients1-3. When such mutants emerge during treatment, famciclovir and other drugs (adefovir, tenofovir) can be used as therapeutic alternatives1. Point mutations related with lamivudine resistance occur within the P open reading frame (ORF) of the HBV genome and affect domains B and C of the HBV reverse transcriptase (RT), leading to various aminoacid substitutions (fig. 1). Aminoacid transitions in positions 528 (L528M) and 552 (M552I, M552V) of the HBV DNA polymerase have been associated with lamivudine resistance. Another point mutation, leading to an aminoacid transition in position 555 (V555I), has been associated with famciclovir resistance. These positions change in the various HBV genotypes, so a genotype-independent nomenclature has been proposed for the mutations, starting with the highly-conserved EDWGPCDEHG motif at the beginning of the HBV RT domain4. The unified denominations are as follows: rtL180M (previously L528M), rtM204I (M552I), rtM204V (M552V) and rtV207I (V555I).
Figure 1. Aminoacid transitions in the HBV DNA polymerase conferring resistance to lamivudine (rtL180M, rtM204V/l) or famciclovir (rtV207I). Such transitions arise from point mutations within the B or C conserved domains from the reverse transcriptase region of the P gene.
Once mutations are produced, selection of resistant mutants occurs under pressure of the drug: wild-type strains are progressively eliminated by the treatment and the viral population gradually becomes enriched with the resistant mutants. This selection reduces the efficacy of the treatment and may be a reason for stopping it. When lamivudine is discontinued, replication of residual wild-type strains is again possible, and this sometimes leads to a rapid increase in viral replication, with sudden episodes of hepatocyte destruction that may produce severe liver necrosis and death5,6. Since the replicative competence of lamivudine-resistant HBV mutants is usually lower than that of the wild-type strains, it is recommended to maintain the treatment for some time after mutant selection has taken place, in order to avoid these serious complications.
Substitution of wild-type strains by drug-resistant mutants takes place over several months of treatment and seems to be more frequent when specific HBV genotypes are involved in the chronic infection7,8. Nonetheless, cases of early emergence of mutants have been described, suggesting that mutations conferring lamivudine resistance were already present in the liver of these patients before starting therapy9. This would mean that these mutant strains circulate among the population, so that a certain proportion of HBV carriers from a given geographical area might have any of them. The emergence of lamivudine-resistant mutants seems to be more frequent in treated patients carrying genotype A HBV strains7. Thus, the prevalence of naturally occurring resistant strains is expected to be higher in countries such as Spain, where genotype A is the most prevalent among the general population. Research aimed at searching for these naturally occurring variants among untreated carriers has not yet been reported from European countries, but a single case of acute, primary HBV infection involving one of these variants has been found in France10. Data from Japan suggest that the prevalence of lamivudine-resistant strains among naive HBV carriers in that country could be as high as 28%11, although the series of carriers studied was limited.
The presence of a drug-resistant HBV variant before treatment could be a reason for selecting an alternative drug for therapy. Thus, routine testing for mutations involved in antiviral drug resistance might be useful for candidate patients. To judge whether this activity would be appropriate, however, prior knowledge of the prevalence of these variants among the HBV carrier population is required. This retrospective study was performed to obtain preliminary data on the prevalence of lamivudine-resistant HBV mutants among Spanish carriers.
Methods
From January 2001 to May 2002, serum samples from 588 HBsAg carriers were sent to a Spanish reference laboratory for detection and quantification of HBV DNA. Samples were tested by both quantitative molecular hybridization and nested-polymerase chain reaction tests (see below); HBV DNA-positive samples from 229 patients were identified, and 79 patients were randomly selected for the study. After performing the tests for detection of drug-resistant mutants, additional samples from some of these patients, obtained before January 2001, were recovered and included in the study.
HBV DNA was quantified by molecular hybridization, using a commercial assay (Digene Hybrid Capture II, HBV DNA Test, Digene Corp., Gaithersburg, MD, USA) and a fragment of the viral genome was amplified by a nested-polymerase chain reaction (n-PCR) in-house developed test, targeted on the P region of the HBV genome. Outer primers HBPr134 and HBPr135 (59 -TGC TGC TAT GCC TCA TCT TC-39 and 59 -CA(A/G) AGA CAA AAG AAA ATT GG-39, respectively) were used in the first reaction to obtain a fragment that was amplified again in a second reaction using nested primers HBPr75 and HBPr94 (59 -CAA GGT ATG TTG CCC GTT TGT CC-39 and 59 -GGT A(A/T)A AAG GGA CTC A(A/C)G ATG-39, respectively)12. A final 341 base-pair fragment, corresponding to aminoacids 465 to 562 of the HBV DNA polymerase gene, was obtained and studied by agarose gel electrophoresis. Since the nested primers were biotinilated, the final amplification product was biotin-labeled, enabling direct testing for the presence of point mutations related with lamivudine or famciclovir resistance by a commercial line probe assay test (INNO-LiPA HBV DR, Innogenetics N.V., Ghent, Belgium)12. The performance of this assay for the detection of the mutations studied as compared to sequence analysis has been found satisfactory13. HBV DNA tests were performed when samples arrived at the laboratory, whereas LiPA assays were done retrospectively on biotin-labeled amplification products stored at 20 °C.
After identifying samples with significant mutations, the corresponding clinical records were reviewed and the clinician responsible for the management of each patient studied was contacted, in order to confirm the data regarding the patient's status with respect to lamivudine or famciclovir treatment at the time of sampling.
Results
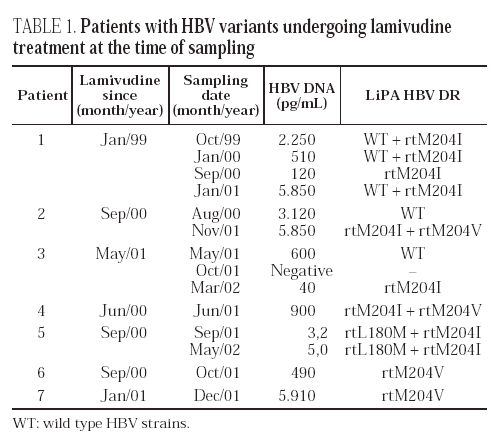
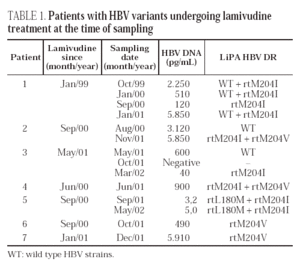
Point mutations related with drug resistance were detected in ten patients (tables 1 and 2). The rtM204I mutant was the most frequent (6 cases), either alone or associated with rtM204V or rtL180M. Single rtM204V or rtV207I mutants were found in two cases, respectively. The rtL180M mutant was found together with rtM204I in one case. Wild-type strains were associated with mutant strains in three patients.
Seven of the ten patients had a record of lamivudine therapy. Mixtures of mutants in a single position (rtM204V + rtM204I) or in two positions (rtL180M + rtM204I) were found in three of them, and single mutants (rtM204I or rtM204V) were detected in the remaining four (table 1). All these mutations would predict resistance to lamivudine. Follow-up samples, taken over a period of 14 months, were available from four patients. On the initial sample, before or at the beginning of therapy, wild type sequences alone were found in two of them, and a mixture of wild type and rtM204I mutant was found in Patient 1. The wild-type strains cleared later on and emerged again at the end of follow-up. This patient had human immunodeficiency virus (HIV) coinfection and received highly active antiretroviral therapy (HAART) together with lamivudine treatment. The patient's compliance with therapy was poor; lamivudine treatment was interrupted during August and September 1999, and started again in October. Compliance with therapy continued to be poor and finally, both HAART and lamivudine treatment were considered unsuccessful and were withdrawn in January 2002. All patients were still undergoing treatment at the time of follow-up sampling.
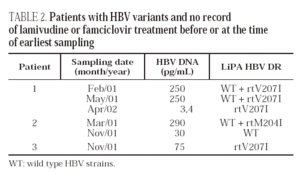
There was no record of treatment with lamivudine or famciclovir in the remaining three patients, 3,8% of the 79 carriers studied. We consider that these cases are likely to reflect natural infections by strains resistant to lamivudine (one case) or famciclovir (two cases) among Spanish HBV carriers. All three patients were men, 32 to 42 years old. None of them were HIV coinfected, a fact that makes unnoticed prior treatment with lamivudine unlikely. The patients had the rtM204I or rtV207I mutation; variant mixtures or strains with multiple mutations were not found (table 2). Wild-type strains together with the drug-resistant variants (rtM204I or rtV207I) were found in two cases. Serum clearance of the wild-type strains apparently occurred in Patient 1, nearly one year after they were last detected. The patient had liver cirrhosis and was diagnosed with chronic hepatitis B in 1992. In Patient 2, the rtM204I variant was not detected on follow-up testing. Clearance of the variant was observed to coincide with exacerbation of liver disease, with increased aminotransferase levels, anti-HBc IgM production, and a decrease in viremia from 98 to 30 pg/mL of viral DNA. No follow-up samples could be obtained from Patient 3.
Discussion
Data supporting the natural occurrence of lamivudine-resistant strains among the population of chronic HBV carriers have been reported recently from Japan and France10,11. Such natural occurrence might also explain the early emergence of resistant mutants observed in South Korea among some patients receiving lamivudine therapy9. On the basis of these observations, routine testing for lamivudine-resistant variants might be useful in chronic HBV carriers scheduled for antiviral therapy, particularly in countries where genotype A strains are prevalent7,8. Since famciclovir is often used as replacement therapy in cases of emergence of lamivudine-resistant variants, research into mutations conferring primary resistance to this alternative drug would also be of interest. In order to assess the appropriateness of optimizing routine testing for HBV drug resistance in a given country, the prevalence of naturally occurring variants among randomly selected, chronic HBV carriers should be investigated.
The results from the present study indicate that HBV variants associated with drug resistance circulate in the Spanish population and suggest a prevalence among chronic HBV Spanish carriers of nearly 4%. Since the replication fitness of these variants is low, it is likely that these infections result from direct transmission from an external source rather than from natural emergence during chronic infection. The 4% incidence found in this study is much lower than the 28% reported from Japan11, but could be significant enough to support the performance of prospective, population-based studies to establish the true prevalence of these variants in Spain and to assess the need for routine screening for lamivudine resistant mutants prior to therapy.
Lamivudine therapy was approved for the treatment of chronic hepatitis B in Spain in 2001. Before receiving this approval, it was used only in very selected cases, except in the case of HIV-infected patients. Therefore, it seems unlikely that the finding of HBV strains with mutations conferring resistance to lamivudine or famciclovir among samples from three HIV-negative patients lacking records of prior antiviral therapy after exhaustive investigation would respond to former, unnoticed treatment with these drugs. It is much more likely to reflect the circulation of these variants within the general population.
Mutations in the YMDD motif from the HBV RT have been shown to produce other effects besides the emergence of lamivudine resistance. Association of the rtL180M mutation with changes in position 204, either rtM204I or rtM204V, restores the replicative competence of rtM204I/V mutants and induces resistance to other antiviral drugs that may be useful in treating chronic hepatitis B12. Changes in the "a" determinant of the HBsAg have also been reported in association with various mutations in the YMDD motif13. In vitro experiments have shown that these changes enhance the replication of resistant mutants in the presence of the drug13. It is possible that modifications in such an important region of the HBsAg may lead to other relevant effects. Thus, testing for these mutations might provide further interesting information on the viral strains infecting a patient, in addition to the specific information about lamivudine resistance.
Routine testing for HBV drug resistance requires robust, reliable molecular assays. The INNO-LiPA HBV DR reverse hybridization test14 used in the present study has correlated well with sequence analysis for the detection and identification of YMDD mutations15. We have found it to be a user-friendly, clean method that can be almost fully automated using the AUTO-LiPA unit. Our experience over the last decade with routine use of LiPA technology for genotyping hepatitis C virus strains has shown that obtaining genome fragments by nested PCR improves the quality of the results, probably because of the high concentration of target material required to obtain clean hybridization patterns. The use of biotinilated primers for the diagnostic PCR nested reaction provides a material that can be safely stored at -20 °C and directly tested by LiPA when indicated, without a significant increase in cost.
Acknowledgements
The authors thank Dr. Campos (Hospital General, Soria), Dr. Casal (Hospital de la Princesa, Madrid), Dr. Casas (Hospital de Alcorcón, Madrid), Dr. Frau (Hospital Santa Bárbara, Ciudad Real), Dr. Infantes (Hospital La Inmaculada, Almería), Dr. Pardo (Hospital General, Castellón) and Dr. Zozaya (Hospital de Navarra, Pamplona) for providing information on their patients with lamivudine-resistant HBV mutants, as well as J.A. López, P. García and I. Parera for their excellent technical assistance.