SARS-CoV-2 is a new pathogen that has emerged in Hubei province, China, on December 2019 and was declared pandemic on March 2020 by the World Health Organization (WHO). Coronaviruses are monopartite, single-strand RNA, positive sense, capped, and virions are enveloped.1 The persistence of SARS-CoV-2 on surfaces varies considerably based on the type of material. Stability is higher on plastic and stainless steel than on copper and cardboard2 and its persistence has also been established on high-touch surfaces in laboratories such as phones and keyboards.3 Nasopharyngeal swab is the recommended sample to detect SARS-CoV-2 but sampling requires trained staff with personal protective equipment, the procedure is uncomfortable for the patients, and may induce sneezing with the consequent risk of aerosol generation. Preliminary studies to detect viral genome on N95 masks have been performed4 including SARS-CoV-2.5 In Spain, the use of surgical masks is mandatory in almost all its territory. To our knowledge, there are no studies addressing this issue with surgical masks.
In this report, we show that SARS-CoV-2 RNA can be detected on surgical masks worn by patients for several hours and we propose that this may be an alternative non-invasive method to detect the presence of the virus.
We selected 4 patients that came to the Emergency ward in our tertiary care hospital with a positive nasopharyngeal sample for SARS-CoV-2. All of their clinical and demographic characteristics were recovered by clinical records. To process the masks we cut out an area of approximately 10cm×17cm covering the nostrils and mouth excluding the outer layer in order to avoid possible false positive results due to external contamination. To inactivate viral particles we used guanidine isothiocyanate; approximately 3mL to cover the mask sample, then, vortexed for a minute and finally we performed RNA extraction and the PCR following the same protocol used routinely in our laboratory for respiratory samples. For the extraction we use the MagMAX Express 96, Magnetic Particle Processor (Applied Biosystems) following the manufacturer recommendations. All samples were tested using the TaqMan 2019 nCoV Assay Kit v1 (Thermo Fisher Scientific Inc. Franklin, MA, USA).
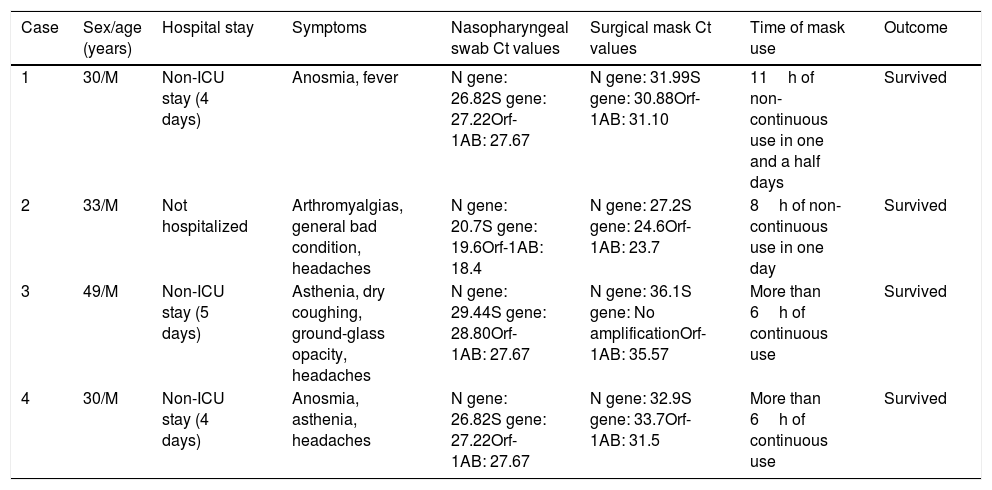
Four patients, all of them men (age range 30–49 years), came to the Emergency ward presenting mild symptoms such as anosmia, asthenia, general bad condition, fever, and headaches. In the X-ray only one patient had ground-glass opacity. Three patients were admitted to the Internal Medicine department due to haematological alterations. All of the patients survived and were discharged after a hospitalization of an average of four days. PCR for SARS-CoV-2 was performed in all patients in nasopharyngeal swabs at their arrival and all three genes amplified (Table 1). When analyzed, all surgical masks also amplified for the three genes, except one, with an average of five Ct values higher than those in the nasopharyngeal swabs (Table 1). The average time of use of the surgical masks was 8h in one and a half day.
Clinical and demographic characteristics of four patients with COVID-19 and their Ct values of nasopharyngeal swabs and surgical masks.
| Case | Sex/age (years) | Hospital stay | Symptoms | Nasopharyngeal swab Ct values | Surgical mask Ct values | Time of mask use | Outcome |
|---|---|---|---|---|---|---|---|
| 1 | 30/M | Non-ICU stay (4 days) | Anosmia, fever | N gene: 26.82S gene: 27.22Orf-1AB: 27.67 | N gene: 31.99S gene: 30.88Orf-1AB: 31.10 | 11h of non-continuous use in one and a half days | Survived |
| 2 | 33/M | Not hospitalized | Arthromyalgias, general bad condition, headaches | N gene: 20.7S gene: 19.6Orf-1AB: 18.4 | N gene: 27.2S gene: 24.6Orf-1AB: 23.7 | 8h of non-continuous use in one day | Survived |
| 3 | 49/M | Non-ICU stay (5 days) | Asthenia, dry coughing, ground-glass opacity, headaches | N gene: 29.44S gene: 28.80Orf-1AB: 27.67 | N gene: 36.1S gene: No amplificationOrf-1AB: 35.57 | More than 6h of continuous use | Survived |
| 4 | 30/M | Non-ICU stay (4 days) | Anosmia, asthenia, headaches | N gene: 26.82S gene: 27.22Orf-1AB: 27.67 | N gene: 32.9S gene: 33.7Orf-1AB: 31.5 | More than 6h of continuous use | Survived |
Rapid and reliable SARS-CoV-2 diagnosis has become a priority in order to control outbreaks and stop the transmission chain. Obtaining nasopharyngeal swabs is an invasive procedure and can be complicated in patients such as children and the elderly.5 Furthermore, it can generate aerosols with increased risk for healthcare workers. In our proof of concept tests, we have proven that RNA material can be detected on surgical masks material. The main advantages of using masks to detect viral RNA are that these are a non-invasive, easy to obtain, that might be particularly useful in certain uncooperative population groups, sampling does not require trained staff, and samples might be used for screening, medical follow-up and also for the detection of other respiratory pathogens5 as William et al. demonstrated for Mycobacterium tuberculosis on mask samples.6 On the other hand, the main limitations of this approach are the heterogeneity between nasopharyngeal swabs and surgical masks, the variability of viral excretion in different types of patients, the larger processing time of the mask sample, more time-consuming than processing the nasopharyngeal sample, less possibility of automation, and that the ideal preservation conditions (avoiding light exposure, refrigerating the mask in a hermetic sheath) can be complex for the patient.5 In our patients the differences in Ct values between the nasopharyngeal swabs and surgical masks could be explained by the discontinued time of use of the mask, lack of ideal conservation conditions of the sample and the rapid clearance of the viral load associated with early clinical improvement in all of our patients. Even though this method seems like a promising non-invasive approach to detect SARS-CoV-2 RNA, more studies are needed to confirm its utility.
FundingThe authors declare no financial support.
Conflicts of interestNone.
The authors declare no financial support.
María Dolores Montero-Vega, María Pilar Romero, Silvia García-Bujalance, Emilio Cendejas-Bueno, Carlos Toro-Rueda, Guillermo Ruiz-Carrascoso, MI Quiles-Melero, Fernando Lázaro Perona, J Mingorance, Iker Falces-Romero, Almudena Gutierrez-Arroyo, I Bloise, B Gómez-Arroyo, Patricia González-Donapetry, Gladys Virginia Guedez-López, Paloma García-Clemente, María Gracia Liras Hernandez, Consuelo García-Sanchez, Miguel Sánchez-Castellano, Sol San José-Villar, Cancelliere-Fernández, Rosario María Torres Santos-Olmo, J García-Rodriguez.