Extended-spectrum β-lactamases (ESBLs) are increasingly prevalent in Enterobacter spp., posing a challenge to the treatment of infections caused by this microorganism. The purpose of this retrospective study was to evaluate the prevalence, risk factors, and clinical outcomes of inpatients with bacteremia caused by ESBL and non ESBL-producing Enterobacter spp. in a tertiary hospital over the period 2004–2008.
MethodsThe presence of blaCTX-M, blaTEM, blaSHV, and blaPER genes was detected by polymerase chain reaction (PCR) and nucleotide sequence analysis. Genetic similarity between strains was defined by pulsed-field gel electrophoresis (PFGE).
ResultsEnterobacter spp. was identified in 205 of 4907 of the patients who had positive blood cultures during hospitalization. Of those cases, 41 (20%) were ESBL-producing Enterobacter spp. Nosocomial pneumonia was the main source of bacteremia caused by ESBL-producing Enterobacter spp. The presence of this microorganism was associated with longer hospital stays. The ESBL genes detected were: CTX-M-2 (23), CTX-M-59 (10), CTX-M-15 (1), SHV-12 (5), and PER-2 (2). While Enterobacter aerogenes strains showed mainly a clonal profile, Enterobacter cloacae strains were polyclonal.
ConclusionAlthough no difference in clinical outcomes was observed between patients with infections by ESBL-producing and non-ESBL-producing strains, the detection of ESBL in Enterobacter spp. resulted in the change of antimicrobials in 75% of cases, having important implications in the decision-making regarding adequate antimicrobial therapy.
Las β-lactamasas de espectro extendido (BLEE) son cada vez más frecuentes en Enterobacter spp., lo que representa un desafío para el tratamiento de infecciones causadas por este microorganismo. El propósito de este estudio fue evaluar los factores de riesgo y los resultados clínicos de pacientes ingresados con bacteriemia causada por Enterobacter spp. productores de BLEE en un hospital terciario durante los años 2004-2008.
MétodosLa presencia de los genes blaCTX-M, blaTEM, blaSHV, e blaPER se detectó mediante la reacción en cadena de la polimerasa (PCR) y análisis de la secuencia de nucleótidos. La similitud genética entre las cepas fue definida por electroforesis en gel de campo pulsado (PFGE).
ResultadosEnterobacter spp. fue identificado en 205 pacientes de un total de 4.907 que tenían cultivos positivos de sangre durante la hospitalización. De esos 205 casos, 41 (20%) eran Enterobacter spp. productores de BLEE. La neumonía nosocomial fue la principal fuente de bacteriemia causada por Enterobacter spp. productores de BLEE. La presencia de este microorganismo se asoció con una mayor estancia hospitalaria. Las BLEE detectadas fueron: CTX-M-2 (23), CTX-M-59 (10), CTX-M-15 (1), SHV-12 (5) y PER-2 (2). Mientras que las cepas de Enterobacter aerogenes presentaron un perfil principalmente clonal, E. cloacae fueron policlonales.
ConclusionesSi bien no fueron observadas diferencias en los resultados clínicos entre los pacientes con infecciones causadas por cepas productoras de BLEE y no productoras de BLEE, la detección de BLEE en Enterobacter spp., resultó en el cambio de los antimicrobianos en el 75% de los casos, habiendo implicaciones importantes en la toma de las decisiones con respecto a la terapia antimicrobiana adecuada.
Enterobacter spp. has been recognized as nosocomial pathogens, mainly affecting patients in the intensive care unit (ICU). E. cloacae and E. aerogenes are the most common species found in human infections. The frequency of bacteremia has increased in the last decade, and the rate of infection is approximately 1/1000 admissions in university hospitals or tertiary-care centers.1 In general, the mortality associated with bacteremia caused by Enterobacter spp. is as high as bacteremia caused by other enteric bacilli, with mean rates of 20–35%.1
β-Lactam resistance in Enterobacter spp. challenges the treatment of infections caused by this organism and is associated with unfavorable clinical outcomes.2 The main resistance mechanism in these organisms is the expression of chromosomally encoded AmpC cephalosporinase,1 leading to treatment failure when third-generation cephalosporins are used.2 Moreover, the frequency of extended-spectrum β-lactamase (ESBL) expression is increasing in these species as an important cause of broad-spectrum cephalosporin resistance.2
While AmpC cephalosporinase is chromosomally encoded, ESBLs are mediated by transferable plasmids. Therefore, detection of ESBL genes is recommended for the adoption of control measures to prevent spread of resistance.3
Several reports have described the high mortality, time, and cost of hospitalization as well as delays in choosing the appropriate antibiotic therapy associated with infections caused by Klebsiella pneumoniae, Escherichia coli, and Proteus mirabilis ESBL-producing microorganisms.4–6 However, studies evaluating the effects of ESBL production in Enterobacter spp. are rare. One report compared the clinical significance of ESBLs produced in Enterobacter spp., Citrobacter spp., Serratia spp., and Morganella morganii and found no differences in the outcome of bacteremia between those microorganisms.7 However, further clinical studies are hindered by the difficulty of detecting ESBLs in species that also express genes encoding inducible AmpC, since the latter may mask the detection of the former.8
This study evaluated the prevalence, risk factors, and clinical outcomes of bacteremia caused by ESBL- and non-ESBL-producing Enterobacter spp. strains.
Materials and methodsStudy settingThis study was conducted at the Clinics Hospital of Federal University of Paraná (HC/UFPR), a 643-bed teaching hospital located in Curitiba, Brazil, over a 5-year period (January 2004–December 2008). Over the past 10 years, HC/UFPR has had an average of 18,699 admissions per year, including 159 ICU admissions, per year. The Clinics Hospital offers a broad range of complex care including bone marrow transplants, liver transplants, cardiovascular surgery, chemotherapy, and other high-risk procedures.
The study was approved by the Institutional Review Board of the HC/UFPR (project number 2288.182/2010-07). All patients received an individual identification number, and their names were not disclosed to maintain confidentiality.
Study design and bacterial strainsA retrospective comparative study was performed to assess risk factors and clinical outcomes of nosocomial infection by ESBL-producing Enterobacter spp. vs. non-ESBL-producing Enterobacter spp. strains. All patients included in this study developed bacteremia after 48h of hospitalization and yielded positive blood culture for Enterobacter spp. (either ESBL-producing or non-producing strains). One isolate per patient was included in the study. The total number of patients with positive blood culture collected during the period of study was considered to assess prevalence.
Clinical data were obtained from medical records of the patients. The following clinical data were evaluated: age and sex, underlying disease (cancer, digestive tract diseases, respiratory diseases, heart diseases, central nervous system diseases, or kidney disease), invasive procedures (central venous catheter, mechanical ventilation, tracheotomy, surgery, transplant, or urinary catheter), ward and/or intensive care unit (ICU) admission, use of antimicrobial agents, immunosuppressant or chemotherapy prior to culture, primary focus of bacteremia, length of hospital stay (days), before and after blood culture results (with ESBL or non-ESBL producing Enterobacter spp.), changes in antimicrobial therapy after physicians were informed of the ESBL results, and clinical outcome (death, discharge, or related death).
DefinitionsPatients with ESBL-producing Enterobacter spp. were defined as “group 1” and patients with non-ESBL-producing Enterobacter spp. were defined as “group 2”. Death was considered related to infection when the cause of death was septic shock and the last microorganism isolated from blood cultures was identified as Enterobacter spp. Antimicrobial therapy with carbapenems for ESBL-producing Enterobacter spp., and carbapenem or fourth-generation cephalosporins for non-ESBL Enterobacter spp. was considered appropriate therapy. Third-generation cephalosporins were considered inadequate therapies for both groups, and other antibiotics were deemed appropriate or not according to the antimicrobial susceptibility test.
Microbiology studiesBlood cultures were performed with the BactAlert® system (bioMérieux, Hazelwood, MO). The Vitek system (bioMérieux) was used for species identification (ID 32 GN card) and antimicrobial sensitivity tests (AST N105 card). Isolates showing reduced sensitivity to cephalosporins were tested (ceftriaxone, ceftazidime, cefotaxime, or aztreonam), according to the Clinical Laboratory Standard Institute (CLSI) breakpoints for ESBL screening in Klebsiella spp. and E. coli,9 were tested for the presence of ESBL-encoding genes.
Molecular detection of ESBL genesAll isolates of Enterobacter spp. with a positive phenotypic screening test were tested for the presence of blaCTX-M, blaTEM, blaSHV, and blaPER genes by polymerase chain reaction (PCR) as previously described.10–12 Positive controls K. pneumoniae A30397 (blaCTX-M-2), E. coli E27 (blaTEM-9), and K. pneumoniae A32048-3 (blaSHV-5), and the negative control E. coli ATCC 25922 were run simultaneously. Samples that were PCR-positive for blaCTX-M were submitted to a second PCR using the primers: blaCTX-M-1 5′-TGTTAGGAAGTGTGCCGCTG-3′; 5′-GACGGCTTTCTGCCTTAGGTTG-3′ and blaCTX-M-2 5′-ATGTTAACGGTGATGGCGAC-3′; 5′-GCATCAGAAACCGTGGGTTAC-3′. Amplicons from all isolates were purified using a GFX-TM PCR purification kit (Amersham Bioscience, NJ, USA) and sequenced using the same set of primers. The nucleotide sequences were identified using a BigDye terminator v3.1 Cycle Sequencing kit and an automated DNA capillary sequencer (ABI PRISM 3700 DNA Analyzer; PE Applied Biosystems, Hitachi), analyzed with ChromasPro version 1.33 (Technelysium Pty LTDA), and compared with the GenBank database sequences using the BLAST tool (http://www.ncbi.nih.gov/BLAST). Isolates with ESBL genes were classified as ESBL-producing.
Pulsed-field gel electrophoresis (PFGE)DNA of all ESBL-producing strains was extracted as described previously13 and digested with XbaI (10U; Fermentas, Maryland, USA) at 37°C. Electrophoresis was performed on a CHEF-DR III (Bio-Rad Laboratories, USA) instrument for 24h at 6V/cm and 12°C and pulse times from 5 to 30s. Gels were analyzed with the Gel-Pro Analyzer 4.0 and NTSYS 2.02 software. Clusters of possibly related isolates were identified using the Dice similarity coefficient and unweighted pair-group method with arithmetic averages (UPGMA). Identical isolates were assigned the same capital letter and different pulsed-field gel electrophoresis (PFGE) profiles were named with different capital letters. Isolates with more than 80% similarity were assigned as a subtype and given the same capital letter as the major type, followed by an Arabic number (e.g., A1, A2, A3, and A4).
Statistical analysesEpidemiological data were recorded and analyzed using Epi Info version 3.5.1. and Statistics v.8.0, using the following statistical parameters: Kruskal–Wallis test for nonparametric continuous variables and chi-square (degree of freedom=1) or Fisher's exact test (expected value less than 5) for categorical variables. A logistic regression for multivariate analyses was included for variables with P<0.05 in univariate analyses. The results were considered statistically significant when the P value was less than 0.05.
ResultsDuring the study period (2004–2008), 4907 patients had positive blood cultures, and Enterobacter spp. were isolated from 205 (4.1%) patients, showing an increasing prevalence of 2.7% in 2004 to 5.8% in 2008 (data not shown). The Enterobacter spp. species found were E. cloacae (109 cases, 53.2%), E. aerogenes (91, 44.4%), and E. gergoviae (5, 2.4%).
Out of the 205 Enterobacter spp. isolated, 41 (20%) were ESBL-producing (20 were E. cloacae and 21 were E. aerogenes) while 164 (78%) were non-ESBL-producing Enterobacter spp.
Patient age ranged between 15 days and 97 years old. We compared the ESBL- and non-ESBL groups and their distribution according to the following age strata <18 (pediatric, n=22), 18–64 (adults, n=119) and ≥65 (elderly, n=64); ESBL-producing strains were observed in 22.7% of pediatric patients, 21% of adult patients and 17.2% of elderly patients. There was no significance difference between groups. There was also no gender difference between ESBL- and non-ESBL-producing groups.
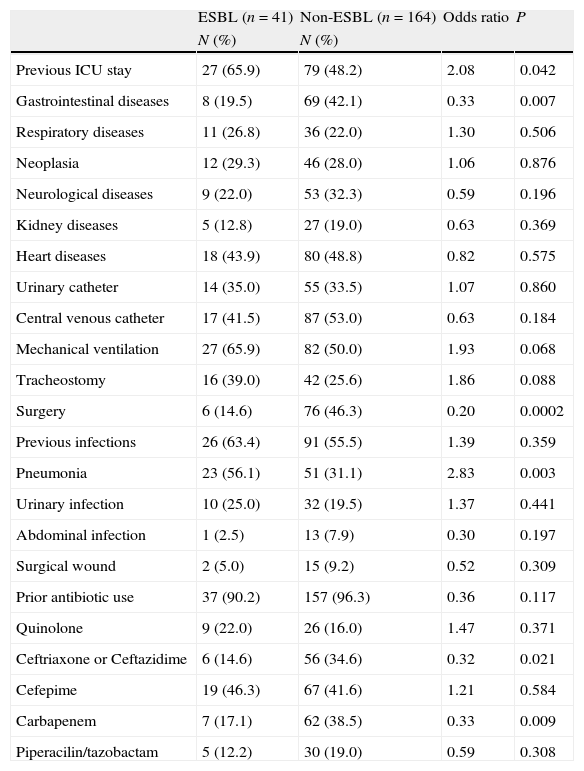
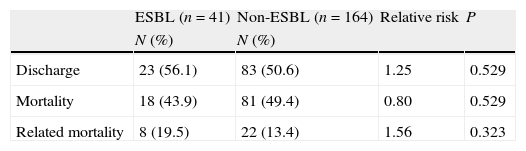
ESBL- and non-ESBL-producing strains showed similar distributions between different hospital wards, except in the ICU where the frequency of ESBL-producing strains was higher. Similarly, the length of hospital stay before infection was similar between the 2 groups, varying only in ICU, where a longer stay was a significant risk factor for ESBL-producing Enterobacter spp. infection (Table 1).
Risk factors of ESBL- and non-ESBL-producing Enterobacter spp. bacteremia.
| ESBL (n=41) | Non-ESBL (n=164) | Odds ratio | P | |
| N (%) | N (%) | |||
| Previous ICU stay | 27 (65.9) | 79 (48.2) | 2.08 | 0.042 |
| Gastrointestinal diseases | 8 (19.5) | 69 (42.1) | 0.33 | 0.007 |
| Respiratory diseases | 11 (26.8) | 36 (22.0) | 1.30 | 0.506 |
| Neoplasia | 12 (29.3) | 46 (28.0) | 1.06 | 0.876 |
| Neurological diseases | 9 (22.0) | 53 (32.3) | 0.59 | 0.196 |
| Kidney diseases | 5 (12.8) | 27 (19.0) | 0.63 | 0.369 |
| Heart diseases | 18 (43.9) | 80 (48.8) | 0.82 | 0.575 |
| Urinary catheter | 14 (35.0) | 55 (33.5) | 1.07 | 0.860 |
| Central venous catheter | 17 (41.5) | 87 (53.0) | 0.63 | 0.184 |
| Mechanical ventilation | 27 (65.9) | 82 (50.0) | 1.93 | 0.068 |
| Tracheostomy | 16 (39.0) | 42 (25.6) | 1.86 | 0.088 |
| Surgery | 6 (14.6) | 76 (46.3) | 0.20 | 0.0002 |
| Previous infections | 26 (63.4) | 91 (55.5) | 1.39 | 0.359 |
| Pneumonia | 23 (56.1) | 51 (31.1) | 2.83 | 0.003 |
| Urinary infection | 10 (25.0) | 32 (19.5) | 1.37 | 0.441 |
| Abdominal infection | 1 (2.5) | 13 (7.9) | 0.30 | 0.197 |
| Surgical wound | 2 (5.0) | 15 (9.2) | 0.52 | 0.309 |
| Prior antibiotic use | 37 (90.2) | 157 (96.3) | 0.36 | 0.117 |
| Quinolone | 9 (22.0) | 26 (16.0) | 1.47 | 0.371 |
| Ceftriaxone or Ceftazidime | 6 (14.6) | 56 (34.6) | 0.32 | 0.021 |
| Cefepime | 19 (46.3) | 67 (41.6) | 1.21 | 0.584 |
| Carbapenem | 7 (17.1) | 62 (38.5) | 0.33 | 0.009 |
| Piperacilin/tazobactam | 5 (12.2) | 30 (19.0) | 0.59 | 0.308 |
ICU=intensive care unit; n=number of patients.
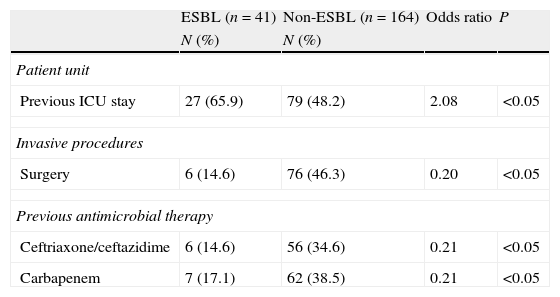
Among all underlying diseases and co-morbidities evaluated, none represented independent risk factors for infection by ESBL-producing strains (Tables 1 and 2). Moreover, invasive procedure during hospitalization was not a risk factor for infection by ESBL-producing Enterobacter spp. On the other hand, the use of carbapenem and third-generation cephalosporins during hospitalization proved to be a risk factor for infections by non-ESBL-producing Enterobacter spp. (Tables 1 and 2).
Multivariate analysis of the risk factors of ESBL- and non-ESBL-producing Enterobacter spp. bacteremia.
| ESBL (n=41) | Non-ESBL (n=164) | Odds ratio | P | |
| N (%) | N (%) | |||
| Patient unit | ||||
| Previous ICU stay | 27 (65.9) | 79 (48.2) | 2.08 | <0.05 |
| Invasive procedures | ||||
| Surgery | 6 (14.6) | 76 (46.3) | 0.20 | <0.05 |
| Previous antimicrobial therapy | ||||
| Ceftriaxone/ceftazidime | 6 (14.6) | 56 (34.6) | 0.21 | <0.05 |
| Carbapenem | 7 (17.1) | 62 (38.5) | 0.21 | <0.05 |
ICU=intensive care unit; n: number of patients.
Regarding the clinical outcome, the mortality among patients with bacteremia by ESBL-producing Enterobacter spp. was 43.9%; among patients with bacteremia by non-ESBL-producing Enterobacter spp., mortality was 49.4%. ESBL- and non-ESBL-producing Enterobacter spp.-related mortality was 19.5% and 13.4%, with no significant difference between groups. The patients infected with ESBL-producing strains had a longer hospital stay after the culture results than patients with non-ESBL-producing Enterobacter spp. bacteremia. The mean hospital stay in this study was 8 days in the ICU and 7 days in other wards. During the course of this study, bacteremia caused by Enterobacter spp. increased the mean length of hospital stay to 15 days and ESBL-producing Enterobacter spp. increased the mean duration of hospitalization to 22 days (Table 3).
Patients infected with ESBL-producing strains that were using empirically ceftriaxone or ceftazidime were considered to have received inappropriate therapy, and totaled 85.4% of the study population. The microbiological diagnostic results led to a change in antimicrobial therapy in 75.6% (31/41) of patients. In most of these cases (97%), the exchange occurred from cephalosporins to carbapenems. On the other hand, ineffective empirical treatment (third generation cephalosporin) was not a risk factor for death or related death in patients with bacteremia in both groups.
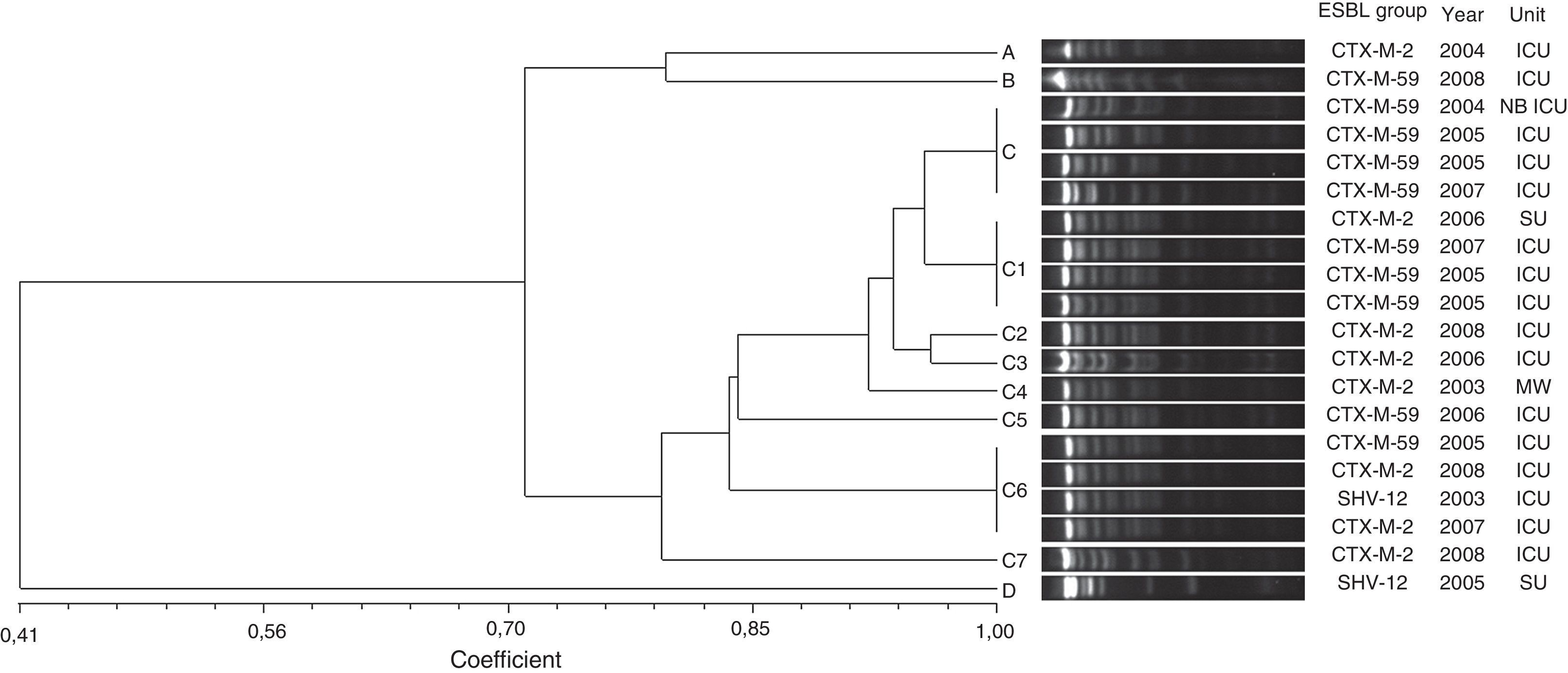
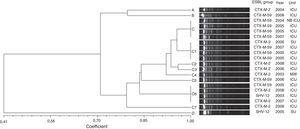
Three different groups of β-lactamases were identified from the total number of ESBL-producing strains: CTX-M (34 isolates), SHV (5 isolates), and PER (2 isolates). CTX-M-2 was the most prevalent type, accounting for 83% of all ESBL-producing strains, including 8 isolates of E. aerogenes and 15 isolates of E. cloacae. CTX-M-59 was detected in 10 isolates of E. aerogenes and CTX-M-15 in 1 isolate of E. cloacae. SHV-12 enzymes were found in 3 E. cloacae and 2 E. aerogenes. PER-2 was found in 2 E. cloacae strains.The high degree of genetic similarity of the E. aerogenes isolates identified in this study is reflected in the results of the PFGE profiling analysis. Out of eleven distinct PFGE types, a PFGE group with at least 80% similarity was identified (group C) (Fig. 1). This group included 85% (17/20) of the isolates and was divided in eight subgroups (C, C1, C2, C3, C4, C5, C6 and C7). For the subgroups C, C1 and C6, clones (100% identity on the basis of PFGE analysis) with four isolates were found. The other PFGE types (A, B and D) showed less than 80% similarity and represented unrelated isolates. CTX-M-59 and CTX-M-2-producing E. aerogenes were frequently found. ESBL groups comprised ten isolates of CTX-M-59 (50%), eight of CTX-M-2 (40%), and two of SHV-12 (10%).
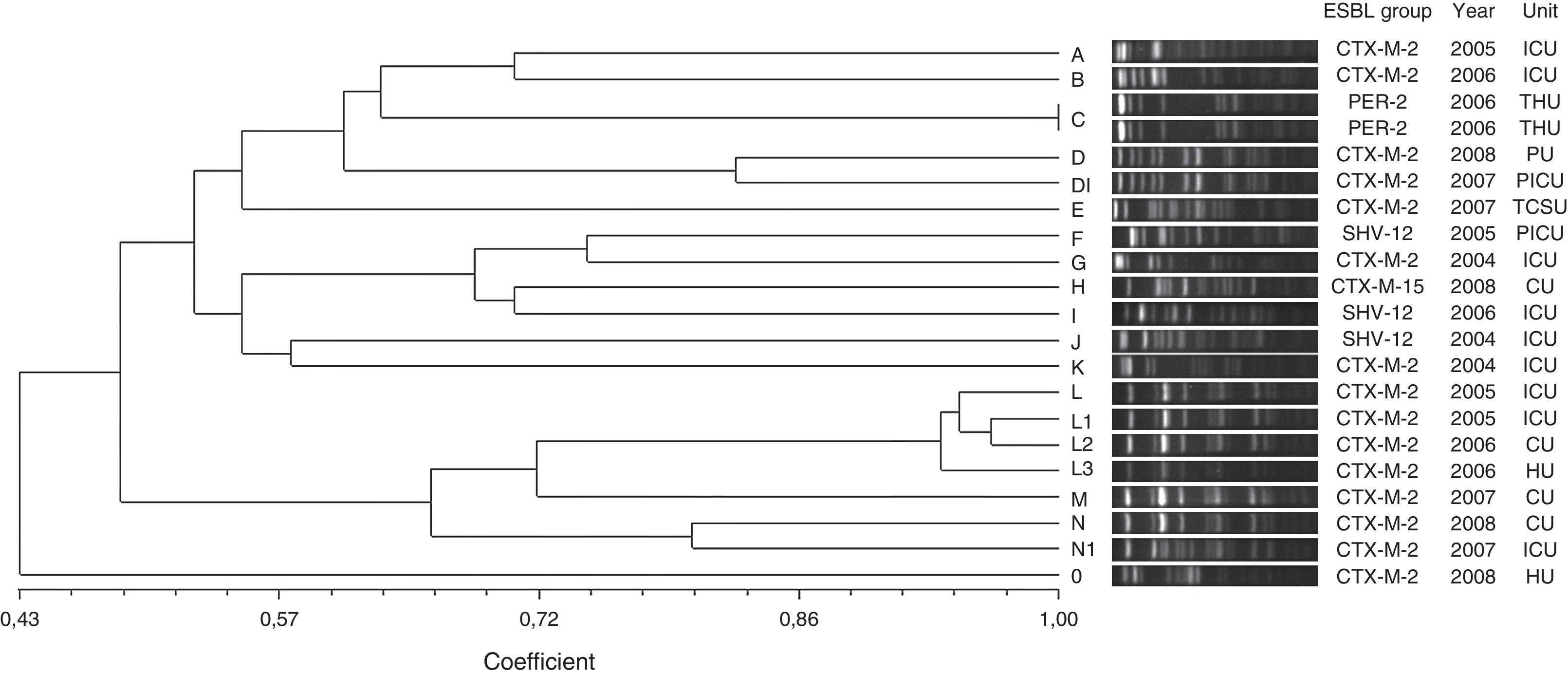
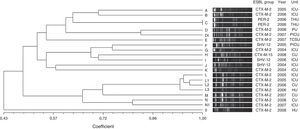
Twenty distinct PFGE types were identified in E. cloacae isolates on the basis of dendrogram analysis (Fig. 2). Two PFGE groups possessed above 80% similarity, group D that contained two isolates and group L that clustered four isolates. Furthermore, these groups included only CTX-M-2-producing E. cloacae. The other PFGE types showed to be unrelated isolates (<80% similarity), and also included CTX-M-2-producing E. cloacae, besides SHV-12, PER-2 and CTX-M-15-producing E. cloacae. Only a clonal group was observed (group C) that comprised two PER-2-producing E. cloacae. This ESBL group has never been described in previous Brazilian studies.
Genetic similarity among ESBL-producing Enterobacter cloacae by PFGE and ESBL type, unit, and year of microorganism isolation. Legend: HU=hematology unit; CU=chemotherapy unit; ICU=intensive care unit; CTCV= Thoracic and Cardiovascular Surgery unit; THU=transplant hepatic unit; P ICU=pediatric intensive care unit; PU=pediatric unit.
The emergence of bacteremia caused by Enterobacter spp. has been described in recent years. In this study, Enterobacter spp. was isolated in 4.3% of patients with bacteremia in a teaching hospital in southern Brazil during the period 2004–2008, with an increased frequency (2.7–5.8%) of isolation over the 5-year study period. A previous study showed similar results,14 and another reported an average of 2.2% (range, 1.4–3.4%).15 These results highlight Enterobacter spp. as a pathogen frequently present in nosocomial bacteremia.
The prevalence of E. cloacae, followed by E. aerogenes, and less participation from other species of this genus as agents of bacteremia, is consistent with the results obtained in other studies.1,15
ESBL production has been found with increased frequency in Enterobacter spp. The frequency of ESBL among Enterobacter spp. in this study was 20%. A study conducted in Korea found 43% of ESBL-producing Enterobacter spp. isolated from blood cultures16; another study conducted in Hong Kong found a frequency of 6.5%.2 These data show that prevalence of ESBL in these species varies, and can be high in some countries.
Risk factors for infection by ESBL-producing strains include previous antibiotic therapy, severity of disease, cross transmission,17 longer hospital stays,17 advanced age, and chronic diseases,15 among others. When bacteremia caused by ESBL- and non-ESBL-producing Enterobacter spp. was evaluated in this study, there were no major differences in the risk factors for infections between the 2 groups. These findings suggest that ESBL has less influence on the selection of resistant organisms in this species, probably because Enterobacter spp. harbored chromosomal AmpC, which confers resistance during therapy with third-generation cephalosporins.
In this study, pneumonia was the main source of bacteremia by ESBL-producing strains, suggesting that ESBL strains have a hospital origin. Moreover, ESBL-producing strains presented mainly a clonal profile, and in 74% of cases, pneumonia was associated with mechanical ventilation. Likewise, ICU stay was relevant for the acquisition of ESBL-producing Enterobacter spp. Furthermore, clonality of E. aerogenes isolated from ICU patients during the 5-year study suggested the hospital origin of the strains.
Studies have associated ESBL production with higher mortality, longer hospital stay, delay in appropriate therapy, and higher treatment costs. Most of these studies compared E. coli, Klebsiella spp., and Proteus mirabilis ESBL- and non-ESBL-producing strains.5,6,15 The present study shows a similar risk of mortality in the group of patients harboring ESBL- and non-ESBL-producing strains. This finding may be explained by the high resistance of Enterobacter spp. to antibiotics, even in the absence of ESBL production. However, ESBL-producing infectious agents increased hospital stay, which typically leads to higher costs of patient care. Costs were not directly assessed, but they are higher because the average length of stay was 7 days longer in patients with ESBL-producing Enterobacter spp. than in patients infected with non-ESBL-producing strains.
The main cause of mortality in infections by E. coli and Klebsiella ESBL-producing strains is the delay in administration of appropriate antibiotic therapy.5,15,18 These data were not evaluated in infections by ESBL-producing Enterobacter spp. in this study.
Beyond the epidemiological significance of ESBL detection, there is also the therapeutic importance. During this study, ESBL detection was used to adjust therapy in 75% of patients with ESBL-producing Enterobacter spp. bacteremia. This action could have affected the association between ESBLs and outcome, suggesting that if ESBL was not detected, mortality might have been even greater.
CTX-M is a type of ESBL widely prevalent in the world, mainly in South America.19 It was also the predominant type identified in this study. PER-2 represents another important type of ESBL, which is prevalent in South America,12 but has never been reported in Brazil and was found in 2 isolates from this study. Similarly, the SHV-12 type, which is commonly found in the USA7 but not in Brazil, was detected in 5 strains.
Cross transmission was more prevalent in E. aerogenes than in E. cloacae. The same type of ESBL was detected in strains not genetically related and different ESBLs were detected in genetically related strains, indicating that transfer of plasmids is probably the most important mechanism of resistance dissemination by ESBLs in this study. ESBL detection in Enterobacter spp. is important to prevent increased frequency of ESBL in Enterobacter spp. as well as in other species of Gram-negative bacilli present in the hospital environment; moreover, the plasmid that carries ESBL genes often carries multiple genes for resistance to other antimicrobials.3
Enterobacter spp. is a pathogen with increasing importance in bacteremia. ESBL production is an important mechanism of resistance to broad-spectrum cephalosporins, reducing the therapeutic options for infection by this microorganism. The predominant type of ESBL was CTX-M, with minor participation of other types such as SHV and PER. The PFGE typing showed the occurrence of clonal and non-clonal spread of the strains, suggesting that the ESBL genes have been horizontally disseminated. ESBL detection in Enterobacter spp could help to control the spread of this kind of resistance. Moreover, the ESBL production had not contributed to mortality but increased the length of hospital stay.
FundingThis study received no funding.
Conflict of interestAuthors have no conflict of interest to declare.