To the Editor: Human papillomavirus (HPV) infection is one of the most common sexually transmitted diseases. Certain HPV genotypes, the so-called high-risk genotypes (HR), have an established role in cervical carcinogenesis. Detection of HR genotypes in cervical samples is an important measure for preventing cancer of the cervix and, together with Papanicolaou smears, is the cornerstone of cervical cancer screening. Currently, two useful assays are available for detecting HPV in cervical samples, hybrid capture (HC) and polymerase chain reaction (PCR). PCR has several advantages over HC, but also some problems; mainly, the test is time-consuming, experience in molecular assays is needed, and cross-contamination is common. A validated, reliable, fast, easy to perform, semior fully-automated assay would be desirable for HPV detection and genotyping, particularly in laboratories that handle a large number of samples. Genotyping methods are important for improving our understanding of the progression of infection to cervical cancer, since individual risk stratification, the prevalence and persistence of infection, and the onset of new infections can be determined by genotyping1. The development of HPV vaccine also depends on typing2.
Stevens et al recently described an automated extraction system, Roche MagNA Pure LC, as an alternative method for specimen processing before using the linear array HPV test (LA) for detecting and genotyping HPV3. The authors recommend the use and validation of automated nucleic acid purification platforms to simplify the assay's extraction step. The Cobas AmpliPrep (CA) (Roche Diagnostics, S.L., Barcelona, Spain) instrument is used for quantification of hepatitis B, hepatitis C, HIV, CMV and other viruses in clinical samples in many clinical microbiology laboratories. Our study sought to compare automated HPV nucleic acid extraction by the CA with the manual AmpliLute (MA) (Roche Diagnostics, S.L., Barcelona, Spain) protocol, a commercially available liquid media extraction kit.
DNA was extracted from 100 cervical brush samples (in PreservCyt medium) and isolated using two different procedures: 1) a 250 μl aliquot was extracted with the AmpliLute Liquid Media Extraction (EXTRN) kit according to the manufacturer's instructions; and 2) a 800 μl aliquot added to 250 μl ATL (EXTRN kit) was extracted using the automatic nucleic acid extraction method of the CA, using a procedure that involved modification of the CS3 cassette of the Total Nucleic Acid Isolation (TNAI) kit (Roche Diagnostics, S.L., Barcelona, Spain). AVE buffer (EXTRN kit) was used instead of elution buffer in the TNAI EB bottle and CAR (EXTRN kit) and diluent were added to the empty GPV vial. Then, all samples were analyzed by LA assay for HPV genotyping by automated system using the recommended protocol, except that a 100-μl aliquot of the CA denatured amplicons was used. The staff carrying out the study had not been specifically trained in molecular techniques. One positive and one negative control were processed with each run of up to 22 specimens. The internal control included in the kit was β-globin. An additional primer pair targets the human β-globin gene, which was isolated concurrently. All 100 samples analyzed generated positive high and low β-globin results (two visible bands for each strip) using the two protocols, thus demonstrating adequate specimen integrity, DNA extraction, and amplification4.
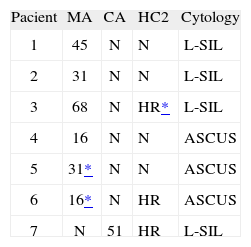
High-risk HPV types were detected by both manual and automated protocols in 45/100 cervical samples; 48/100 samples tested negative, a nearly perfect concordance level (kappa = 0.86 ± 0.099). Manual AmpliLute detected six additional HR-HPV-positive samples (2 HPV-16, 2 HPV-31, 1 HPV-45, and 1 HPV-68) and CA revealed one additional positive result (HPV-51). An alternative test, Hybrid Capture II (HC2) (Digene, Gaithersburg, USA), was used to analyze discrepant samples. Four HR-HPV were negative and three positive, one of them with low signal intensity. Table 1 lists the results of these 7 discrepant samples with all the methods used (MA, CA and HC2), as well as the cytology data5. Some cross-contamination cannot be ruled out in the MA results. In fact, cross-contamination was not unexpected considering the manner in which the manual method is performed. Our data show that for detecting HR-HPV in cytology specimens with borderline changes, MA detected slightly more cases and reached a higher analytic sensitivity than CA. Both tests performed similarly with respect to identification of HPV in high-grade disease.
In practical terms, several molecular methods have been described for identifying type-specific HPV genotypes5. Although PCR-based assays have not yet been approved by the U.S. Food and Drug Administration for clinical use, they provide a rapid means of HPV detection and genotyping in clinical practice. The LA HPV genotyping test is a standardized, consistent, rapid method for detecting and typing up to 37 HPV mucosal genotypes6. Nevertheless, MA can be time-consuming and labor-intensive in clinical settings where many cervical samples are processed. In this context, and based on our data, we propose to replace the manual extraction step by the automated method evaluated in our study. This method provides several advantages and, in agreement with Stevens' results3, the two extraction methods demonstrated only slight differences in the outcome, although a small number of samples were analyzed and larger studies would be desirable. The use of an automated system for DNA isolation would greatly improve simplicity, time and labor efficiency, and reduce potential sample cross-contaminations, thus allowing many cervical samples to be processed simultaneously for cervical cancer screening.