MALDI-TOF mass spectrometry is now a routine resource in Clinical Microbiology, because of its speed and reliability in the identification of microorganisms. Its performance in the identification of bacteria and yeasts is perfectly contrasted. The identification of mycobacteria and moulds is more complex, due to the heterogeneity of spectra within each species. The methodology is somewhat more complex, and expanding the size of species libraries, and the number of spectra of each species, will be crucial to achieve greater efficiency. Direct identification from blood cultures has been implemented, since its contribution to the management of severe patients is evident, but its application to other samples is more complex.
Chromogenic media have also contributed to the rapid diagnosis in both bacteria and yeast, since they accelerate the diagnosis, facilitate the detection of mixed cultures and allow rapid diagnosis of resistant species.
La espectrometría de masas MALDI-TOF es ya una herramienta de trabajo rutinaria en Microbiología Clínica, por su rapidez y fiabilidad en la identificación de microorganismos. Sus resultados están perfectamente contrastados en la identificación de bacterias y levaduras. La identificación de micobacterias y hongos filamentosos presenta mayor complejidad, por la mayor heterogeneidad de espectros dentro de cada especie. La metodología es algo más compleja, y la ampliación del número de especies de referencia, y del número de espectros de cada especie, serán cruciales para alcanzar mayor eficacia. La identificación directa a partir de hemocultivos se ha implantado dada su aportación al manejo de pacientes graves, pero su aplicación a otras muestras es más compleja.
Los medios de cultivos cromogénicos han supuesto también una aportación al diagnóstico rápido tanto en bacterias como en levaduras, ya que aceleran el diagnóstico, facilitan la detección de cultivos mixtos y permiten un diagnóstico rápido de especies resistentes.
The introduction of matrix-assisted laser desorption/ionisation time-of-flight (MALDI-TOF) mass spectrometry (MS) represents, in all likelihood, the most important technological change that has occurred in Clinical Microbiology in the last decade. In just a few years it has gone from being a promising novelty to a technology which is fully integrated into daily clinical activity and, in Spain, it is available in the Microbiology Departments of many hospitals.1 This has required mass spectrometers to undergo a significant technological evolution. In this regard, two breakthroughs turned out to be crucial. First, in 1946, WE Stephens designed the time-of-flight (TOF) system, which made it possible to separate different masses. When ions are accelerated in an electrical field and all acquire the same kinetic energy, the speed that they acquire, and therefore the time that they take to cross the vacuum tube, depends on the mass of the ionised molecule, which may be inferred from the time taken in this crossing.
The second milestone was the development of methods that ionised intact proteins which, due to their size, had not been susceptible to ionisation until then, since the high energy required for ionisation ended up altering or destroying the protein itself. In 1987, Köichi Tanaka presented a new method of analysis (soft laser desorption) which transferred to molecules the energy required to ionise them without breaking their fragile chemical bonds. Tanaka was awarded the Nobel Prize in Chemistry in 2002 for this discovery, together with John B. Fenn, for “the development of methods for identification and structural analyses of biological macromolecules”. It was not long before mass spectrometers based on this type of analysis were developed. However, some changes had to be made to the method before MALDI-TOF MS was finally developed. In MALDI-TOF MS, desorption of ions is promoted by a matrix that absorbs laser energy and partially transfers it to the molecules being studied. Although these modifications improved the technique by making it simpler and more sensitive, those responsible for these improvements, Michael Karas and Franz Hillenkamp, were not co-recipients of the Nobel prize. This generated significant controversy. The contributions of these authors enabled macromolecules and biopolymers to begin to be studied using MS. This opened up new fields of application for this technology.
In its nearly 30-year history, MALDI-TOF MS has been used to quantitatively and qualitatively analyse proteins of various origins. Initially, it was applied to previously isolated proteins or small sets of proteins. However, thanks to technical advances in terms of both instrumentation and tools for computerised data analysis, it may now be used to study large groups of proteins.
The possibility of beginning to study complex proteins, not just small peptides, greatly broadened the potential fields of application of MS. One of them, the identification of microorganisms, was quickly applied to clinical practice as the procedure was simplified and the software required to make use of the raw data provided by the spectrometer was improved.
MALDI-TOF mass spectrometry in Clinical MicrobiologyUntil the introduction of MALDI-TOF MS, bacterial identification, even with significant advances such as the creation of miniaturised identification panels and the automation of their inoculation and reading, continued to draw support from the methods developed by traditional bacteriology. Virtually all identification systems continued to be based on the fermentation of sugars and their detection through the change in pH generated, the metabolism of other substrates and the production of different metabolites and enzyme activities that could be detected by chemical methods. All these methods had several limitations:
- •
They required bacterial growth, which in most cases meant an incubation period of at least 16–18h from inoculation to reading.
- •
They showed problems of identification in all those microorganisms with difficulty growing in the liquid media used for inoculation of these panels, as well as in microorganisms with limited biochemical and enzyme activity.
- •
It was necessary to consider the margin of error deriving from the fact that individuals from the same species could react differently to different substrates.
These limitations, while known and assumed, were more obvious when, in view of the discrepancies observed in some studies between MALDI-TOF MS and conventional identification, 16S rRNA sequencing demonstrated that correct identification matched that provided by MALDI-TOF MS in the vast majority of cases.2
In this regard, MALDI-TOF MS has obvious advantages:
- •
Regardless of the potential for identifying microorganisms directly from some samples, which is addressed in another section, even very limited or early plate growth yields reliable identification in a short period of time, thus saving at least those 16–18h of growth in biochemical identification systems.
- •
Analysis of the protein profile of the microorganism on the 2–20kD spectrum, where most ribosomal proteins are located, offers for the vast majority of bacterial species a specific profile, which distinguishes them from all others with a reliability similar to that offered by 16S rRNA sequencing.
To this must be added the fact that the introduction of this technology has greatly expanded the range of genera and species that may be identified reliably with a method likely to be used routinely. This has even led to a re-evaluation of the role as pathogens of microorganisms that, due to the difficulty of identifying them by classic methods, were very likely underdiagnosed.3,4
Anhalt and Fenselau suggested using MS to identify microorganisms as early as 1975.5 The first study demonstrating the efficacy of MALDI-TOF MS for the identification of microorganisms based on complete cells was published twenty years later.6
An article that was likely key to raising awareness of the possibilities of MALDI-TOF MS among specialists involved in the diagnosis of infectious diseases was published in 2009.7 It studied more than 1600 isolates, including Gram-positive and Gram-negative aerobic and anaerobic bacteria, and obtained a rate of correct identifications of 95.4%. Already this article raised a matter that later proved important for the practical utility of this methodology: the availability of sufficiently extensive microorganism databases, both qualitatively (number of genera and species included) and quantitatively (the authors demonstrated that the likelihood of correct identification was higher for those microorganisms for which at least ten different profiles had been entered in the database).
This marked the start of the rapid expansion of the use of this technology in Clinical Microbiology. The first publication in Spain, in 2010, showed a rate of correlation with the conventional methodology, on a species level, of 100% in Gram-positive bacteria and 87.7% in Gram-negative bacteria.8 Since then, the number of publications regarding different aspects of the clinical use of MALDI-TOF MS, especially the identification of microorganisms, has risen exponentially.7–12
Identification of Gram-negative bacteria using MALDI-TOFStudies have demonstrated that its efficacy in the identification of enterobacteria and other Gram-negative bacteria is excellent, including both the most common Gram-negative bacteria in clinical practice and other Gram-negative bacteria that are less common or more complicated to identify with the classic methodology (different species of Yersinia, Aeromonas, Plesiomonas, Brucella, Francisella, Achromobacter, Stenotrophomonas, Burkholderia, etc.). As in other cases, identification problems have almost always been more closely tied to database insufficiencies than to method limitations, as has occurred in some studies with genera such as Ralstonia, Elizabethkingia and Sphingobacterium.13 Initially, some limitations that could have greater clinical importance, such as the inability to distinguish between Escherichia coli and Shigella14 or difficulty distinguishing between Salmonella enterica serovars were raised. More recent studies have suggested that it is possible to overcome these limitations. Computer programmes such as FlexAnalysis and ClinProTools (Bruker Daltonics GmbH, Germany) have identified specific peaks distinguishing between E. coli and Shigella in 90% of cases.15 Other authors have demonstrated that this distinction may be made even without using any additional software by simply increasing the number and variety of profiles of E. coli and Shigella present in the reference database.16 Using this method, the authors correctly identified 60/64 E. coli isolates and 110/116 Shigella isolates.
Everybody who has used MALDI-TOF MS for identification is aware that the usual methodology identifies Salmonella reliably on a genus level, but much less reliably beyond this level. However, publications since 2004 have suggested the existence of specific peaks that could identify serovars more reliably.17 A recent study suggested the existence of peaks that reliably identify the serovar Typhi.18 Since differential elements seem to exist, enlarging the reference database with a higher number of spectra of at least the most common serovars would probably improve the resolution in this specific case.
In other cases, such as differentiation between species of the Enterobacter cloacae complex, the capacity of MALDI-TOF MS is around 80%. This, while not optimal, represents a significant improvement over the conventional methodology.19
Moreover, as already mentioned, the correct identification of microorganisms previously identified incorrectly, such as the genus Raoultella, which is commonly identified by classic methods such as Klebsiella or Enterobacter, is giving rise to a truer vision of their previously underestimated role as pathogens.3
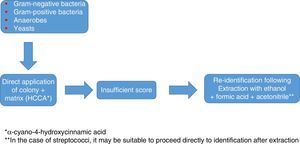
Identification of Gram-positive bacteria using MALDI-TOFThe identification of Gram-positive microorganisms tends to show somewhat lower figures, due in some cases to difficulty lysing the wall and in other cases to the similarity between species. In general, the direct method (where the bacteria are applied directly on the target plate and overlaid with the matrix) tends to be sufficient to achieve good colony-based identification; however, prior extraction with ethanol/formic acid may sometimes be needed to attain optimal identification values. Overall, it yields excellent results in the identification of Staphylococcus aureus and other species and subspecies of both coagulase-producing and non-coagulase-producing staphylococci, as well as enterococci and other less common pathogens such as Micrococcus, Gemella and Rothia. It has also demonstrated its usefulness in the identification of Gram-positive bacilli, including some that are complex to identify using traditional methods (Listeria, Lactobacillus, Corynebacterium, Nocardia, Actinotignum, etc.)20.
In addition, as in the case of Gram-negative bacteria, the systematic use of this technology is changing the consideration as pathogens of some microorganisms, such as Staphylococcus lugdunensis.21 Overall, the data available make it advisable in the case of staphylococci to use, as in any other case, databases offering a higher number of species and a reasonably high number of profiles per species; to employ an extraction method instead of direct application; and to apply the microorganism to the plate in duplicate. Reducing the score required to validate an identification on a species level to values >1.7, rather than the usual values of >2.0, as some authors have proposed,22 may increase the number of identifications on a species level, but at the expense of a drop in specificity, the relevance of which is debatable.
Among Gram-positive bacteria, the streptococci group is probably the one that presents the most problems in terms of identification. While some of the main pathogenic species (Streptococcus pyogenes, Streptococcus agalactiae) are identified with high reliability, Streptococcus pneumoniae (S. pneumoniae) has always been a source of problems, as it is mistaken for other species belonging to the mitis group. The main equipment in use in Spain reliably identifies S. pneumoniae. The most common mistake is erroneous identification of other streptococci from the mitis group, such as S. pneumoniae, especially with the Bruker database.23 Some peaks have been reported as specific to S. pneumoniae; therefore, some authors recommend using them to corroborate these identifications.23 However, the latest Bruker Daltonics reference library (MBT 6903 MSP Library) continues to recommend the use of other tests (optochin, bile solubility) to distinguish between them.
Even within the same group, the quality of the identification of the different species may vary substantially, as occurs within the Streptococcus anginosus group.
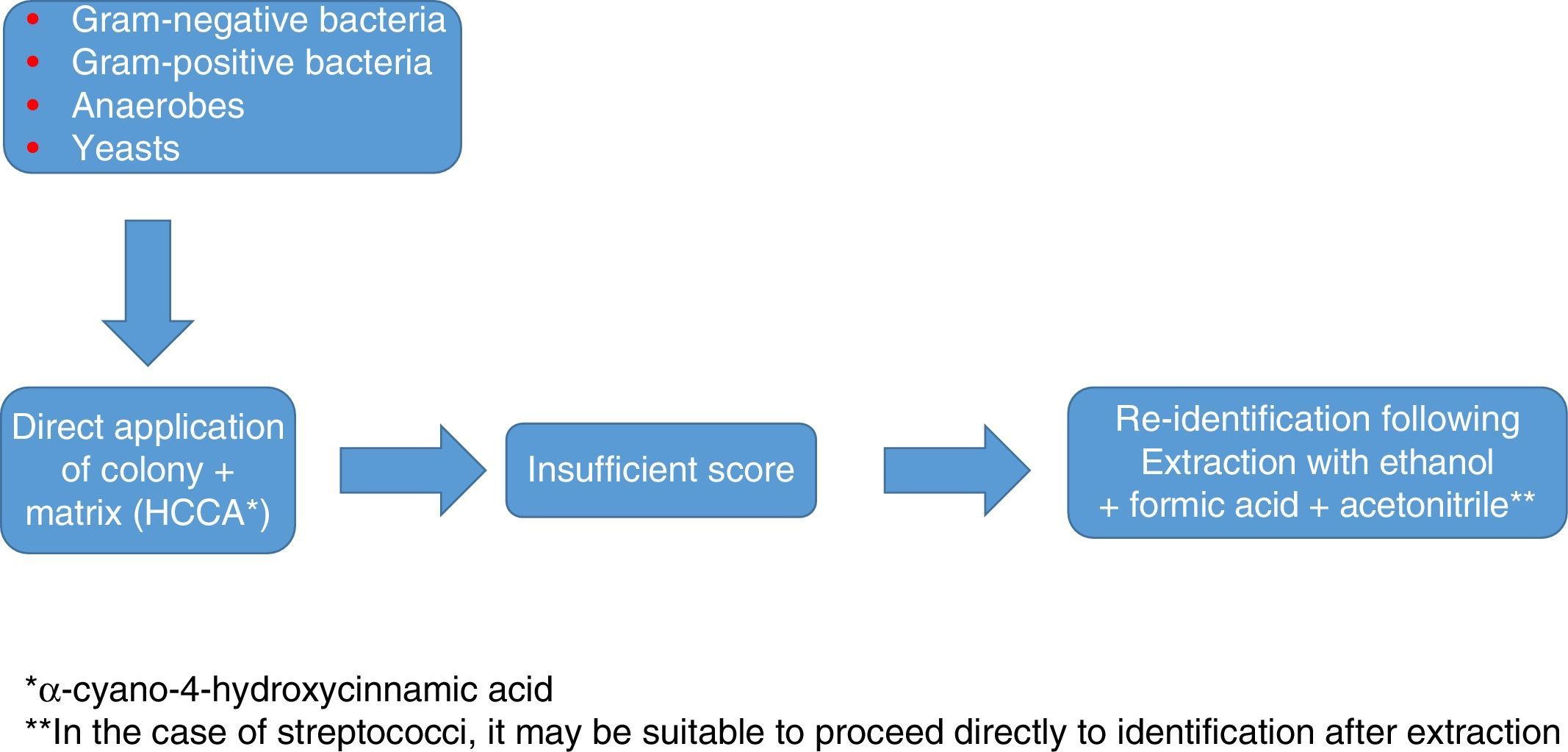
In any case, as with staphylococci, the use of a database that is as extensive and complete as possible, and identification in duplicate with protocols that include extraction, are advisable to optimise results (Fig. 1).
Identification of anaerobic bacteria using MALDI-TOFOverall, MALDI-TOF identifies both Gram-positive and Gram-negative anaerobic bacteria with a high degree of reliability. It correctly identifies the most common species (Bacteroides, Prevotella, Porphyromonas, Fusobacterium, Clostridium, Actinomyces, etc.). Furthermore, the use of MALDI-TOF MS in this field has demonstrated that biochemical identification was much less reliable than previously thought.3 At that point, its combination of simplicity, speed and reliability turned MALDI-TOF MS into the method of choice for the routine identification of anaerobic bacteria in clinical practice. The extraction or non-extraction of the sample does not seem to significantly modify the results. The culture media of origin also does not seem to achieve this. A decisive factor, as in other cases, is the extensiveness of the databases. One of the first studies conducted in Spain3 showed that most errors in identification of anaerobic bacteria were due to the absence of reference profiles of the corresponding species in the database. Earlier studies offered an efficacy in identification of around 75–80%, while more recent studies, with more complete databases or specific databases, have offered figures in excess of 90%. The capacity to identify Clostridium difficile ribotypes, suggested by some authors, has not been definitively demonstrated.
Identification of yeasts and filamentous fungi using MALDI-TOFMALDI-TOF has yielded excellent results in the identification of yeasts from the beginning. It has even distinguished between species that are complicated to differentiate by means of conventional methods, such as the Candida parapsilosis complex. However, its results in relation to filamentous fungi have been much more erratic. Some initial studies showed good results by selectively studying spores; however, this method is impractical in a clinical laboratory, and so most studies have involved joint study of spores and hyphae. In the case of fungi, conventional extraction with formic acid and acetonitrile significantly improves results. Other methods that have been tested, such as the use of glass beads to fragment the walls, do not seem to yield results significantly superior to conventional extraction.
One problem raised by filamentous fungi is the existence of significant differences in the protein profiles obtained, depending on the age of the cultures, even between different subcultures of the same strain.24 The solution to this includes the preparation of more extensive and complex databases featuring profiles of a higher number of strains and cultures of different ages. Some manufacturers recommend extraction following a culture in broth for one day, although this delays results by 24h. A study conducted in 201125 demonstrated that a well-prepared and sufficiently complex database is essential, and may improve identification of filamentous fungi up to figures similar to those obtained with bacteria and yeasts. Unfortunately, the preparation and validation of these databases are complex undertakings that are not within the capabilities of many users, and few of the ones that have been developed are available. Therefore, and also for purposes of standardisation, it is probably most suitable to use manufacturers’ databases and duly extend them.
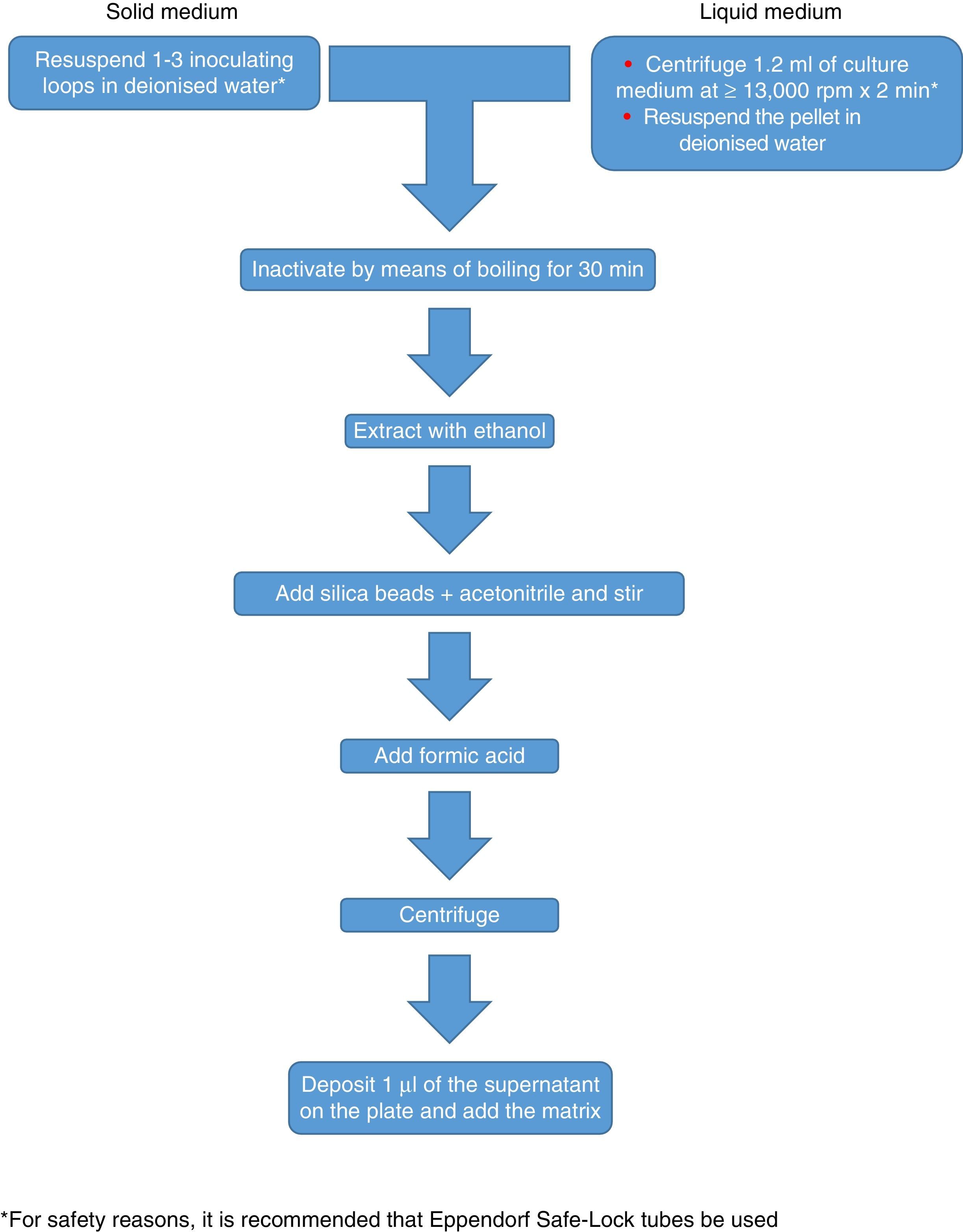
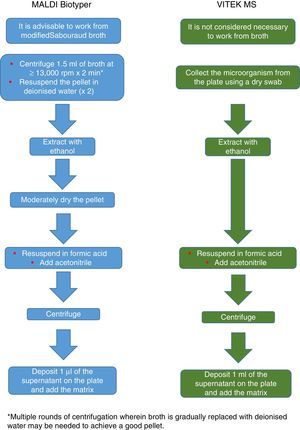
Currently, Bruker Daltonics, whose database of filamentous fungi (version MBT 6903 MSP Library) includes 25 genera and 42 species, recommends culture in a liquid medium for one night, followed by centrifugation, washing and extraction with ethanol, formic acid and acetonitrile (Fig. 2). MS-VITEK (version 3.0) includes 32 genera and 81 species, and SARAMIS (version RUO 4.13) includes a more extensive database with 45 genera and 168 species. bioMérieux does not recommend culture in a liquid medium; instead it recommends a conventional extraction with ethanol, formic acid and acetonitrile (Fig. 2). Nevertheless, a recent study using the methodology recommended by Bruker correctly identified, on a species level, just 72% of isolates,26 and a very recent study with VITEK (version 3.0) correctly identified 66.8% of 318 isolates, due more than anything to deficiencies in the database.27 Therefore, at present, the identification of filamentous fungi using MALDI-TOF MS cannot entirely replace the conventional identification methodology.
Identification of mycobacteria using MALDI-TOFDue to the limitations of traditional methods used to identify mycobacteria, especially with respect to response time, virtually all laboratories have opted for molecular methods of identification. In identifying mycobacteria, MALDI-TOF MS represents an alternative that is as fast as, and less expensive than, molecular techniques. However, it shows peculiarities in several regards. On the one hand, direct analysis methods, which involve strongly concentrating or heavily manipulating microorganisms before inactivating them, are not recommended for biosafety reasons. The use of an extraction method that ensures cell rupture improves the quality of the spectra and increases the safety of the procedure.
Several factors may influence the efficacy of identification of mycobacteria using MALDI-TOF MS. As also demonstrated with fungi, mycobacteria may have very different protein profiles, depending on the age of the cultures. Therefore, it is important for diagnosis to include protein profiles of cultures of different ages for each microorganism in the databases. Mention has also been made of the potential formation of polymers which mask the actual size of the characteristic proteins used for identification. All this renders the identification of mycobacteria a less predictable process as regards results compared to other bacteria, as demonstrated by the heterogeneity of the published data. Significant improvements were made to the libraries in the latest updates. For example, the Biotyper mycobacteria library (version 3.0) now includes 149 species, and VITEK MS (version 3.0) now features 48 more species than the previous version.
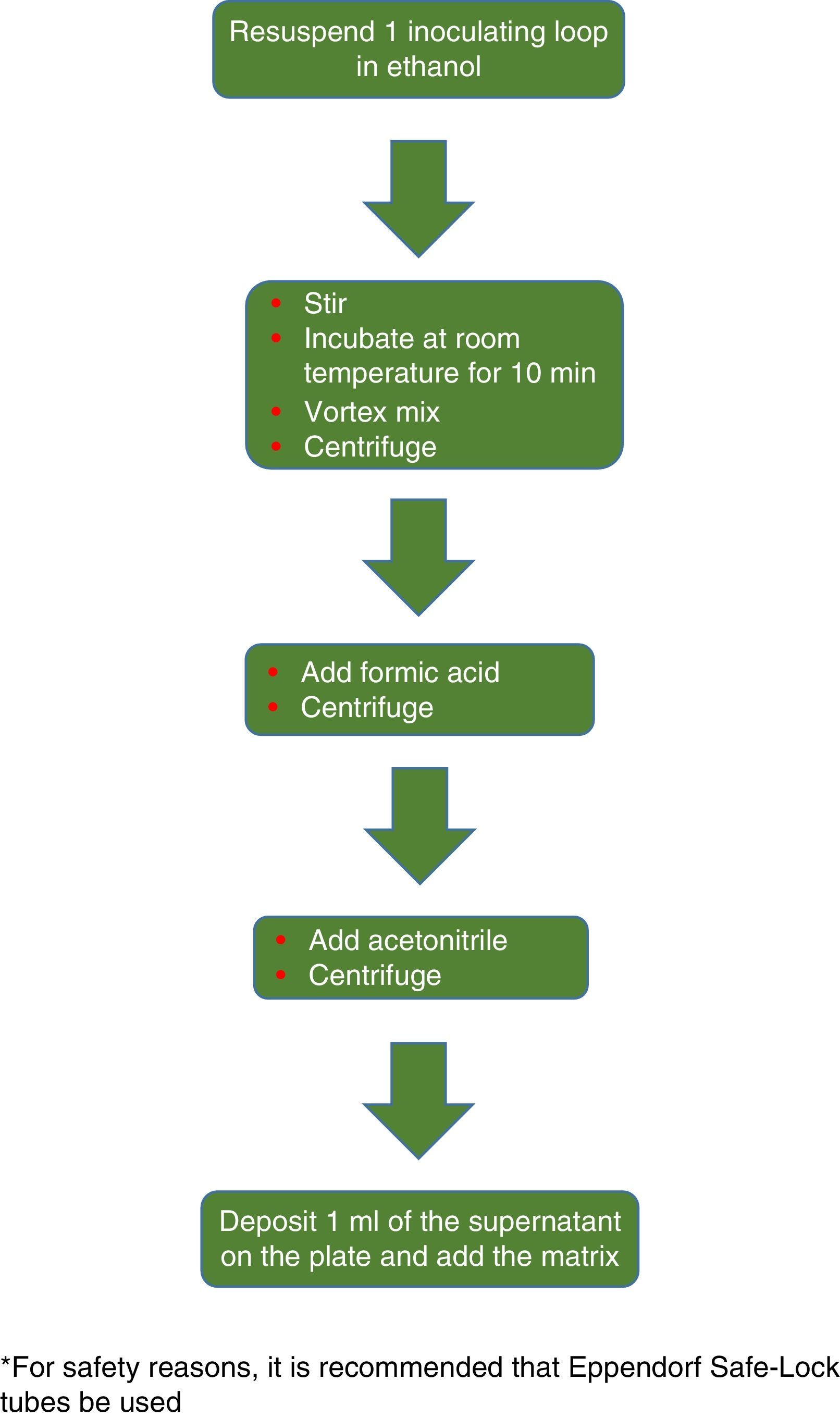
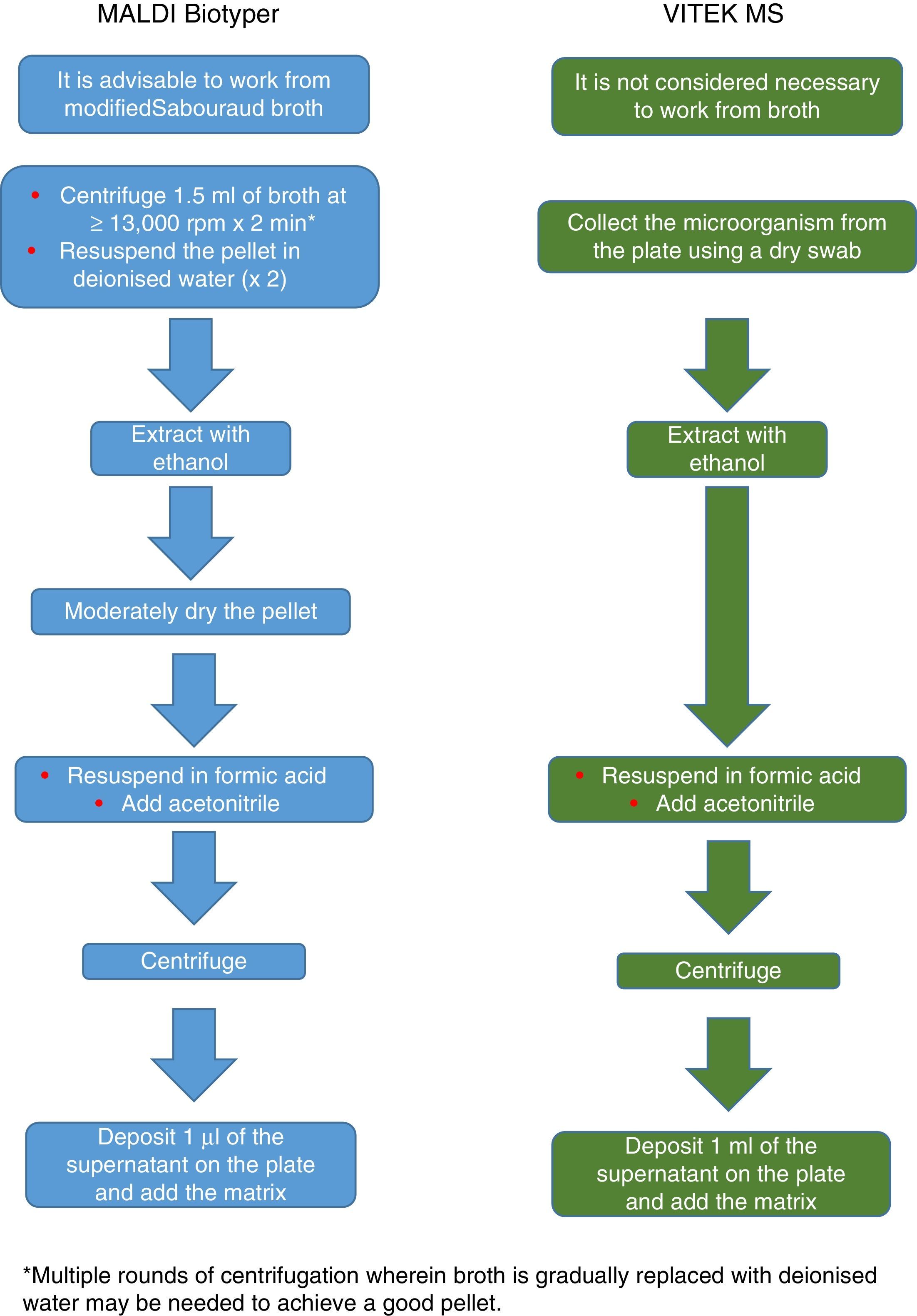
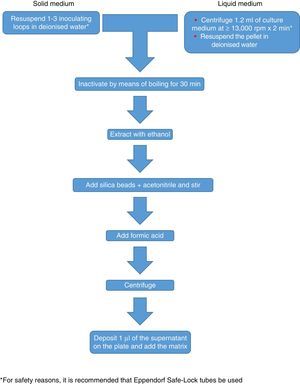
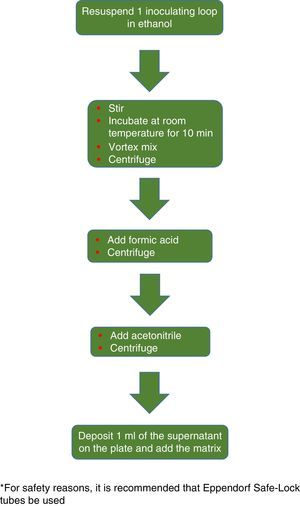
The procedure initially recommended by Bruker included a series of steps (adjustment to a certain optical density, several steps of centrifugation and resuspension) involving a likely unnecessary biological risk. This series of steps has been replaced with a single heat-treatment step. The method currently recommended includes several steps of heating and washing, then a treatment with silica beads and acetonitrile. The samples are then treated with formic acid, applied to the plate, and overlaid with the matrix (Fig. 3). By contrast, the method advocated by bioMérieux is based on initial processing with 70% ethanol and silica beads, following which the sample is extracted with formic acid and acetonitrile, applied to the plate, and overlaid with the matrix (Fig. 4).
A recently conducted study comparing the two methods28 showed that although identification from 7H10 medium tends to be somewhat better than identification from Lowenstein Jensen medium, the differences between the two systems were minor. However, it demonstrated that the sample processing method is critical, and that it is not interchangeable between systems. The samples obtained with the Bruker extraction method yielded better results with the Bruker spectrometer, and those processed with the VITEK method yielded better results both with SARAMIS and with VITEK MS itself, although the difference in this case was smaller. Overall, VITEK MS seemed to be less affected by the extraction method. Regarding efficacy in identification, Bruker Biotyper identified 81.5% of isolates with a score ≥1.8, while SARAMIS and VITEK MS 3.0 identified 85.4% and 89.2%, respectively, with a reliability ≥90%. Using these same criteria, 22.6% of the isolates identified by Bruker Biotyper needed to be extracted more than once to achieve identification. This occurred in 21.5% with SARAMIS and in 14.2% with VITEK MS 3.0.
Regarding other microorganisms with a wall structure close to that of mycobacteria, for some time their identification was believed to be problematic due to the composition of their wall. However, it has been demonstrated that, if a database with sufficient references is available, an extraction method similar to that used with mycobacteria may yield good results.29
All the data referred to above suggest that the spectrum of microorganisms susceptible to identification using MALDI-TOF MS is extraordinarily broad and that, with few exceptions, the limiting factor of the capacity of MALDI-TOF MS almost always lies more in the completeness of the reference databases than in the capacity of the method to obtain reliable profiles of almost any bacterial microorganism susceptible to growing in culture media. In fact, the manufacturing companies seem to be aware of the problem. Thus, the latest reference library published by Bruker Daltonics (MBT 6903 MSP Library) includes 938 new profiles; 17.2% of these are intended to cover new genera and species (specifically, 15 new genera and 96 new species), especially anaerobes and fungi, but 82.8% of these are intended to increase the diversity of profiles within species already included in previous libraries. Similarly, VITEK MS (version 3.0) features 242 new bacterial species, including 48 new species of mycobacteria and 15 new species of Nocardia, and 55 new species of fungi, with an overall average of more than 10 isolates and 25 spectra per species.
Identification of microorganisms, using MALDI-TOF, from a direct sampleFrom the beginning, the potential for identification of microorganisms prior to the growth of colonies has been a very appealing objective. When empirical treatment may be started, it allows for more targeted treatment, reduces the emergence of resistant strains and optimises spending.30 MALDI-TOF MS, applied to a direct sample, has become a very useful tool in serious infections with high rates of morbidity and mortality, such as bacteraemia and fungaemia, even if the direct study of samples has disadvantages that may limit the usefulness of the technique. It is not possible to perform proper identification in samples with low microbial density, since not enough bacterial proteins are present. Something similar occurs with polymicrobial samples, since an aberrant protein spectrum may be generated as a result of the mixture of several profiles, or the microorganism which is in a lower proportion may be completely overlooked. Some software improvements have been developed which may sometimes detect and even separately identify the microorganisms involved; however, they must be carefully studied and probably debugged to be genuinely useful. In most methods that work directly on a sample, given the potential presence of intracellular microorganisms, it is advisable to use methods that include cell lysis.
Not all samples are valid for this type of study. The ideal sample is one extracted from a normally sterile area, which may host high concentrations of microorganism and in which there are no significant limitations with respect to sample volume. Samples taken from areas that are normally colonised (skin, faeces, upper respiratory tract, etc.) will in all likelihood generate aberrant profiles. A technical matter to be taken into account when working directly on a sample is the volume of sample available. In cases such as urine or blood cultures, this does not tend to be a problem. However, in other cases such as cerebrospinal fluid, it may indeed be a problem, and this problem will be all the more significant the lower the bacterial concentration. In addition, samples with a high content of protein whose origin is foreign to the microorganism may also pose problems when they are being interpreted.31
However, due to the advantage that it represents, especially in blood cultures, in speeding up the identification of the microorganism by 24h and more specifically in guiding treatment, the direct use of MS on a sample, especially in these cases, has become widespread.
Direct identification based on urine samplesUrine is one of the samples that best fits the ideal conditions for working with MALDI-TOF MS on a direct sample. It is a sterile fluid under physiological conditions, the bacterial load in infected urine is high in the majority of cases and the limitations with respect to sample volume and acquisition of new samples if necessary are minimal.
Studies have been published with excellent results, with rates of concordance in identification as high as 90–95%32,33 compared to automated systems. Different sample-processing protocols have been reported. All of them agree that it is necessary to perform prior screening (flow cytometry, Gram staining) to distinguish between positive and negative samples, and to limit the use of MS to initially positive samples.34
With slight variations, preparation of samples tends to consist of subjecting them to multiple rounds of centrifugation with deionised water, then performing the conventional MALDI-TOF MS procedure. Modifications have been made by adding SDS 10% to the aliquot of the sample33 to improve protein release, or Tween-8034 to improve the results. The results obtained tend to be better with Gram-negative bacteria than with Gram-positive bacteria and fungi, and the reliability improves greatly when the counts are high (above 105CFU/ml).
A disadvantage of this method is the need to perform the sensitivity study using the conventional methodology. A recent study suggested that, once identification using MALDI-TOF MS has been achieved, the same sediment may be used to make a disk-plate antibiogram with excellent results and thus reduce the length of the full study by 24h.35 Due to the significant organisational change that the introduction of this methodology represents, and the fact that these are infections in which speeding up diagnosis by a few hours is not critical for patient management, diagnosis of urinary infection using MALDI-TOF MS has not become as well established as, for example, diagnosis of urinary infection using blood cultures. Both greater sensitivity and greater standardisation of the methods must be achieved before its introduction in routine clinical practice may be considered.
Direct identification based on blood culturesStarting correct empirical treatment is decisive in the clinical course of bacteraemia. The more accurate and specific the information that can be provided quickly, the higher the likelihood that the treatment started is correct. In this regard, the possibility offered by MALDI-TOF MS of achieving reliable identification shortly after the blood culture becomes positive represents an obvious improvement.
Many blood culture processing protocols have been described (differential centrifugation, cell lysis and protein extraction through chemical methods, separation through gel, etc.). The majority of them are based on lysis and elimination of cell components, either through centrifugation and washing, in which different compounds such as saponin, ammonium chloride and SDS are used, or through the use of tubes with serum separator gel and clot activator. Ultimately, these methods seek to isolate and concentrate microorganisms until a concentration of at least 105–107CFU/ml is achieved, whereupon there are enough proteins to generate suitable profiles in MALDI-TOF MS.
In the case of blood cultures, there is an approved commercial procedure (Sepsityper, Bruker Daltoniks GmbH, Germany) consisting of adding a lysis solution to an aliquot of 1ml of blood culture, then subjecting the sample to multiple washing and centrifugation steps and finally performing a conventional extraction with ethanol and formic acid.36
The results of direct processing of blood cultures are generally good, with percentages of correct identification of 80–90%, although with some slight differences. The identification of Gram-negative bacteria tends to be correct in 90–95% of cases, while in Gram-positive bacteria it is much more heterogeneous, ranging between figures similar to those of Gram-negative bacteria and figures around 50%. Within these, streptococci from the viridans group and non-coagulase-producing staphylococci raise particular problems of reliability.
In initial studies, the results in cases of fungaemia were disappointing. Proper extraction, which is indispensable in this case, yields figures for identification in excess of 90%. These figures are comparable to those obtained in bacteraemia.37 The potential repercussions of certain components of some blood cultures, such as activated charcoal, which may interfere with identification, must be taken into account. In addition, a prolonged blood culture incubation period may have negative repercussions for the spectra obtained. However, since the usual procedure is to perform MS within a few hours of the blood culture becoming positive, and more than 80% of significant positive blood cultures are positive in the first 48h, the clinical impact of this matter does not seem to be important.
When assessing the heterogeneity of the percentages of correct identification in blood cultures, it must be taken into account that there is no uniform criteria for assessing the scores obtained. Some authors have considered correct identification to correspond to values >1.7. Others have decreased this exigency to 1.5, but have included requirements, such as that identification be repeated in the first two or three positions of the list of potential identifications, or that there be a difference in score of at least 0.3 points between the first two options offered by MALDI-TOF.
In general, we are not in favour of reducing the level of exigency in correct identification in blood cultures, since the existence of a higher percentage of “non-identifications” will always represent a lower risk, from a clinical point of view, than the proliferation of incorrect identifications, which could lead to unsuitable treatments.
Regarding the use of commercial processing methods, such as Sepsityper, their results generally resemble those of manual processing methods. Each laboratory must weigh up the greater standardisation and the savings in terms of processing time provided by these methods against the economic savings (around €1/sample) provided by the use of a manual method and set its own priorities.
Overall, MALDI-TOF MS is a fast and reliable method for directly identifying microorganisms in blood cultures. Combining it with methods that detect mechanisms of resistance to certain antimicrobials yields valuable clinical information in a significantly (24–48h) shorter period of time than that required by the conventional methodology. The main point to be elucidated at this time is probably how to incorporate this new activity into Microbiology laboratories in order to optimise the information offered without overloading departments that generally do not exactly have a surplus of technical staff and do have highly variable shift structures.
Direct identification based on other samplesThe number of studies concerning the direct application of MALDI-TOF MS to other biological samples is more limited. As mentioned elsewhere, the usefulness of MALDI-TOF MS in these samples is determined by several factors. On the one hand, the sample should be from a sterile area under physiological conditions and in which the infection, when it appears, tends to be monomicrobial. Moreover, both the bacterial load present in the sample and the sample volume available for study are crucial. This means that many samples other than urine and blood culture pose problems, due to the low volume usually available (CSF, purulent exudates), the low concentration of microorganisms (CSF, peritoneal fluid in patients who undergo peritoneal dialysis) or the presence of significant amounts of non-bacterial proteins which may alter profiles or interfere with ionisation of bacterial proteins (purulent exudates).
Due to the low sensitivity in those products in which the bacterial load is not very high, and the volume of sample available, which is often limited, molecular techniques are generally preferable for rapid diagnosis, even though they are limited in that they are capable of detecting a much lower range of microorganisms in a single test than MALDI-TOF MS. Aetiological diagnoses of bacterial meningitis using direct MALDI-TOF MS on CSF have occasionally been reported, but one study presented at the 21st ECCMID indicated that, out of 183 samples of CSF, 14 were positive using the conventional method, and that out of these samples, none was positive using direct MALDI-TOF MS on a sample.38 This reaffirmed the fact that, from a sensitivity perspective, MALDI-TOF MS cannot compete with molecular techniques. In other sample types, experience is very limited and there are no published protocols or data regarding sensitivity and specificity, and so the clinical use of MALDI-TOF MS is not relevant at present.
Chromogenic mediaSince the development of the first chromogenic medium (CM) more than 30 years ago, chromogenic medium have become very useful tools for the differential isolation of pathogenic microorganisms. At present, there are many different CM marketed for the identification of bacteria and fungi, as well as for the study of some mechanisms of resistance to antimicrobials.
The basis of these media is the inclusion of a chromogenic substrate that, when hydrolysed by a specific enzyme present in the microorganism, gives rise to a colony with a characteristic colouration, thereby enabling its differentiation. They are often also selective media, which inhibit to a greater or lesser extent the growth of other microorganisms and thus promote the detection of the coloured colonies. The main substrates of commercial CM are indole derivatives39 which may be hydrolysed by galactosidases or glucosidases, thereby producing non-toxic derivatives that do not inhibit bacterial growth.
Detection of Gram-positive microorganismsMajor efforts have been dedicated to developing methods to detect methicillin-resistant S. aureus (MRSA), given its significance in nosocomial infection. In this context, the use of CM has gained a great deal of significance for the rapid isolation and identification of this bacterium. There are many CM for the detection of MRSA on the market. A recent study40 compared three of them: chromID MRSA SMART (SMART), chromID MRSA first generation (chromID) and Brilliance MRSA (OX2) for the screening of 1220 samples from hospitalised patients. Detection in these media was corroborated using MALDI-TOF MS, diffusion in agar with cefoxitin disks and commercial PCR for mecA and mecC. Sensitivity at 24h was better for the SMART medium compared to the chromID medium, but there were no significant differences with the OX2 medium. However, the authors indicated that enrichment broth for 24h before inoculation is still needed in any of the media studied for best results.
Xu et al.41 reviewed 5 of the media most commonly used to detect MRSA. In this study, the best results were obtained with Brilliance MRSA 2 (Oxoid Ltd., Thermo Fisher, United States), with a sensitivity of 65.7% and a specificity of 99.8%. The yield of this medium improved if the sample was inoculated in an enrichment broth before it was seeded in the CM, with a sensitivity of 100% and a specificity of 99.1%. The CHROMagar MRSA medium showed a sensitivity of 95% after 24h of incubation which rose to 100% after 48h of incubation. The specificity in both cases was 100%. The BBL CHROMagar MRSA medium includes cefoxitin in its composition, which simplifies interpretation of the results. This medium was studied using MRSA isolates obtained from blood cultures, with a sensitivity and a specificity of 97.6% and 99%, respectively. MRSA Select was evaluated using 652 isolates from blood cultures following 18–24h of incubation at 35°C, with a sensitivity of 99% and a specificity of 98%, which increased to 99% when it was combined with the coagulase test. The chromID MRSA medium, which also includes cefoxitin in its composition, was studied in isolates from wounds and blood cultures. In blood cultures, it showed a sensitivity of 97.8% after 24h, which increased to 100% after 48h, and a specificity of 99.7%, which stayed steady (99.6%) after 48h. In wounds, its sensitivity and specificity were 88.9% and 100% after 24h and 100% in both cases after 48h.
Culture media of this type are complex to compare, since there are many factors, in addition to the culture medium itself, that may influence the results (sample type, inoculum concentration, incubation time, etc.). However, the data for sensitivity and specificity available suggest that any of the above-mentioned media may be routinely used to detect MRSA with high reliability.
Another group of bacteria that has gained importance in nosocomial infection, to the point that the CDC recommend the carrier study at centres with a high prevalence, is vancomycin-resistant enterococci (VRE). The VRE carrier study is conducted using bile esculin azide with vancomycin (BEAV). Its use involves a diagnostic delay of 48h, in addition to the need to perform complementary tests for correct identification. Various CM have been developed to streamline this process. A recent study compared five CM for VRE using 400 faeces samples.42 The media used were InTray Colorex VRE (BioMedDiagnostics, White City, OR), chromID VRE (bioMérieux, Marcyl’Étoile, France), VRESelect (Bio-Rad, Marnes-la-Coquette, France), HardyCHROM VRE (Hardy Diagnostics, Santa Maria, CA) and Spectra VRE (Remel, Lenexa, KS), using BEAV agar and BEAV broth as a reference method. The reading was performed after 24h. The results of the five media showed a sensitivity of 89.9–94.9%, which was higher in all cases than that shown by the BEAV agar (84.9%). The best data for sensitivity and specificity were still provided by BEAV broth, but at the expense of a diagnostic delay of up to 48h compared to CM. The most sensitive CM in this study was chromID VR (94.9%), although there were no statistically significant differences from the other media.
The five CM had similar specificity and capacity for distinguishing between Enterococcus species. The InTray Colorex VR medium had the lowest specificity, with 98.8%, while those of the other four plates were 99.7%. The characteristics of these media in terms of sensitivity and specificity render them advisable for detecting VRE carriers when this is appropriate. They are at least perfectly comparable to BEAV agar and decrease detection time by 24–48h.
Detection of Gram-negative microorganismsIn 1979, Kilian and Blow reported a new culture medium using β-glucuronidase as a substrate to directly detect E. coli in urine cultures. Cost-effectiveness studies indicated that this method represented an economic saving of 46% and a reduction in identification time of 64% compared to conventional methods.39 Since then, many CM have been developed with different substrates for the identification of the main uropathogens. One recently marketed medium is chromID CPS Elite (bioMérieux, Durham, NC), which directly identifies E. coli and presumptively identifies Enterococcus spp., some Enterobacteriaceae and bacteria from the Proteae group, although in these cases identification must be confirmed using biochemical tests or MALDI-TOF MS.
The effectiveness of this medium for detecting uropathogens,43 as well as the time required for diagnosis and the consumables used, were evaluated compared to blood agar and MacConkey agar. The rates of concordance of this medium with the conventional methodology were 88% for clinically significant urine cultures, 74% for urine cultures with non-significant growth, 69% for contaminated urine cultures and 95% for plates without bacterial growth. The main discrepancies were due to growth in blood agar, but not in the CM for Gram-positive bacteria, generally with little clinical significance in urine cultures such as Staphylococcus spp. and Lactobacillus. Regarding the time elapsed between sample seeding and colony identification, the difference was not statistically significant if the two media were compared overall (27.2h on average for the conventional method versus 26.6h for CM); however, the difference was statistically significant in those urine cultures in which E. coli grew in pure culture (27.1h in the conventional method versus 24.4h in chromID, p<0.0001). The authors argued that this improvement in time was due to the ease and reliability of identification given the colour of the colony, which prevented the need for complementary identification methods.
The chromID™ CPS (CPS4) (bioMérieux, St. Laurent, QC) and UriSelect™ 4 (URS4) (Bio-Rad, Montreal, QC) media were evaluated44 on 903 urine samples, also compared to the conventional method of blood agar and MacConkey agar. The rates of concordance with the conventional method were 89.3% and 89.5% for URS4 and CPS4, respectively. If only those samples in which growth was deemed clinically significant were considered, these rates of concordance increased to 93% for the URS4 medium and 93.1% for the CPS4 medium.
In all cases, the authors agreed that the greatest advantage of using CM is the fact that they streamline the identification process as they reduce the need for complementary tests and thus allow an aetiological diagnosis to be made more quickly.
Given the demonstrated usefulness of this diagnostic tool, several groups are designing new CM, some of which have not yet been marketed. These may represent interesting alternatives in the future. A new medium designed45 for the detection of Bacteroides includes in its formulation 3,4-cyclohexanol esculetin-β-d-glucoside, which targets the β-glucosidase activity in Bacteroides and gives rise to the appearance of black colonies. This medium was compared to bile esculin agar in 100 faecal samples. The CM identified Bacteroides fragilis in 34 samples, while bile esculin agar only detected this microorganism in 19 samples. In addition, the CM was much more selective, since growth of species not belonging to the genus Bacteroides was not observed in any case, while growth of microorganisms belonging to other genera (enterobacteria, Clostridium, Candida, etc.) was observed in bile esculin agar on 34 occasions. The authors also studied the capacity of these CM, supplemented with meropenem and metronidazole, to detect the presence of Bacteroides resistant to one or another antimicrobial. They demonstrated that in both cases they are capable of detecting resistant strains with a sensitivity of 100% and a specificity of at least 85%.
Another new medium developed is CHROMagar Yersinia enterocolitica. This medium has an advantage over the cefsulodin–irgasan–novobiocin (CIN) medium in that it has the potential for growth of Y. enterocolitica serovar O3 and Y. pseudotuberculosis strains, whose growth may be inhibited in the CIN medium.46
Detection of yeastsThe first CM developed for fungi, in the early 1990s, had little success in clinical application due to their limited capacity for differentiation. However, the CM for mycology developed in the last 15 years have had much more success, since they enable a quick presumptive identification of different species of yeasts, a better study of mixed cultures and early identification of species associated with resistance to antifungals. As in the previous cases, the media developed are based on the presence of substrates for one or more enzymes (β-N-acetylhexosaminidase, and in some cases, β-glucosidase or phosphatase) which enable quick identification of at least Candida albicans, and in many cases definitive or presumptive identification of Candida glabrata, Candida lusitaniae, Candida kefyr, Candida parapsilosis, Candida tropicalis and Candida krusei. The studies available and the already extensive experience with this type of media have demonstrated a high specificity for identification and a good sensitivity. However, given the variability in terms of colour and morphology that may be seen in isolates from the same species, identification should always be considered presumptive. A recent study on more than 5600 positive clinical samples for yeasts demonstrated that more than 8% were mixed cultures which were much more likely to be detected if CM were used and provided an algorithm combining CM and MALDI-TOF MS as a more suitable identification procedure.47
Conflicts of interestThe authors declare that they have no conflicts of interest.
Please cite this article as: Siller-Ruiz M, Hernández-Egido S, Sánchez-Juanes F, González-Buitrago JM, Muñoz-Bellido JL. Métodos rápidos de identificación de bacterias y hongos. Espectrometría de masas MALDI-TOF, medios cromogénicos. Enferm Infecc Microbiol Clin. 2017;35:303–313.