Several outbreaks of Enterovirus 68 (EV-D68) have recently been reported in the USA and Canada, causing substantial hospitalisation of children with severe respiratory disease. The acute flaccid paralysis detected in the USA and Canada among children with EV-D68 infection has raised concerns about the aetiological role of this EV serotype in severe neurological disease. The circulation of EV-D68 in the general European population seems to be low, but European Centre for Disease Prevention and Control (ECDC) recommends being vigilant to new cases, particularly in severely ill hospitalised patients. In October 2014, enteroviruses were detected in respiratory samples collected from five hospitalised patients, children and adults. Phylogenetic analysis of partial VP1 sequences confirmed that the detected enteroviruses belonged to the D68 serotype, which were also similar to strains reported in USA (2014). However, all five patients developed respiratory symptoms, but only one required ICU admission. None of the patients described had symptoms of neurological disease. Other considerations related to the detection methods used for the diagnosis of respiratory enteroviruses are also discussed. In conclusion, additional evidence has been provided that supports the role of EV-D68 in respiratory infections in hospitalised patients.
En EEUU y Canadá, desde el pasado verano, se han descrito varios brotes causados por EV-D68 afectando a pacientes, principalmente niños, con enfermedad respiratoria grave. En algunos de estos casos la infección por EV-D68 se ha asociado a enfermedad neurológica grave. Aunque en población europea la circulación de este serotipo parece ser baja, el ECDC recuerda la necesidad de reforzar la vigilancia, especialmente en pacientes hospitalizados. En octubre de 2014, en las muestras respiratorias de 5 pacientes ingresados en el Hospital Universitario Vall d’Hebron de Barcelona, se pudieron detectar enterovirus. Estos fueron caracterizados como EV-D68 mediante análisis filogenético de las secuencias parciales codificantes para la proteína viral VP1. Estas secuencias eran además similares a las de las cepas aisladas en los últimos meses en EEUU. Estos 5 pacientes presentaron síntomas respiratorios, pero sólo uno requirió ingreso en la Unidad de Cuidados Intensivos. Sin embargo, ninguno de los pacientes presentó síntomas de enfermedad neurológica. En este trabajo se comentan también consideraciones relacionadas con los métodos de diagnóstico para enterovirus, especialmente para este serotipo. En conclusión, en este trabajo se confirma el posible papel etiológico del EV-D68 en la infección respiratoria del paciente ingresado.
Enterovirus 68 (EV-D68) was first isolated in California in 1962 from cases with bronchiolitis, pneumonia and exacerbation of asthma, and has been rarely reported until 2010.1,2 In the last 5 years the circulation of different EV-D68 strains has been documented worldwide,1–4 and the infection has been associated with mild to severe respiratory disease, being considered as an emerging respiratory pathogen. In addition, a large outbreak is currently occurring in North America (Canada and United States)5 and causing substantial hospitalisation of children with severe respiratory disease, some of them with acute neurologic illness.6 The circulation of EV-D68 in the general population seems to be low, although the risk of EV-D68 transmission in European countries remains moderate. In fact, epidemic clusters of severe disease by EV-D68 have not been still reported. Therefore, European Centre for Disease Prevention and Control (ECDC) reminds of the need to be vigilant to new EV-D68 cases in Europe, particularly in severe hospitalised patients.
In order to accomplish with the enhancement of surveillance tasks led by ECDC to monitor the likely circulation of EV-D68 in European countries, the respiratory specimens from patients at high risk of severe EV-D68 infection attended at a tertiary care university hospital in Barcelona (Spain) were screened. In the present study the first hospitalised EV-D68 cases in Spain in October 2014 were reported.
Materials and methodsFrom 1st September 2014 to 31st October 2014 upper and lower respiratory tract specimens were collected from paediatric and adult patients admitted in our hospital with symptoms of acute respiratory infection (ARI) for laboratory confirmation of respiratory virus infection. Only the patients whose samples were studied by real-time multiplex RT-PCR (Anyplex II RV16 Detection Kit, Seegene, Korea), that allows the detection of the most prevalent respiratory viruses such as human adenovirus (hAdV), influenza A/B virus, human parainfluenza virus (hPIV) 1/2/3/4, rhinovirus (RV), human respiratory syncytial virus A/B, human bocavirus (hBoV) 1/2/3/4, human metapneumovirus, human coronavirus (hCoV) 229E/NL63/OC43 and human enterovirus (EV), were included in the present study. Among this heterogeneous group of patients, all cases with suspicion of viral ARI admitted to the units of Intensive Medicine, Onco-Hematology, Pneumology, Neurology and Neonatology were included to increase the chance to detect EV-D68 associated with severe respiratory disease. The specific EV detection was simultaneously performed by a commercial real-time RT-PCR (SmartEV, Cepheid, USA), not only to enhance EV detection, but also to avoid the likely false results due to the high genetic similarity between rhinoviruses and enterovirus (in particular for EV-D68).1 Institutional Review Board approval was obtained from the Hospital Universitari Vall d’Hebron Clinical Research Ethics Committee. Phylogenetic analysis of the sequences of partial EV VP1 region1 was carried out using MEGA v67 to determine the type of the detected enteroviruses. Sequences from previous molecular epidemiological studies were downloaded from GenBank (GB) Database to be included in the phylogenetic analysis (GB accession numbers are shown in the taxons of the phylogenetic tree) together with recent EV-D68 characterised in The Netherlands and USA.4 The molecular evolutionary models of nucleotide substitution were fitted to the multiple sequence alignment using evolutionary analyses conducted in MEGA v6.7 The phylogenetic tree was constructed using a neighbour-joining distance method as implemented in MEGA v6 with the evolutionary model with lowest Bayesian information Criterion score.7 The topological accuracy of the tree was evaluated by the bootstrap method (1000 replicates). The amino acid composition of partial VP1 amino acid sequences was studied using MEGA v6 relative to the homologous sequence of Fermon Strain with accession number AY426531.
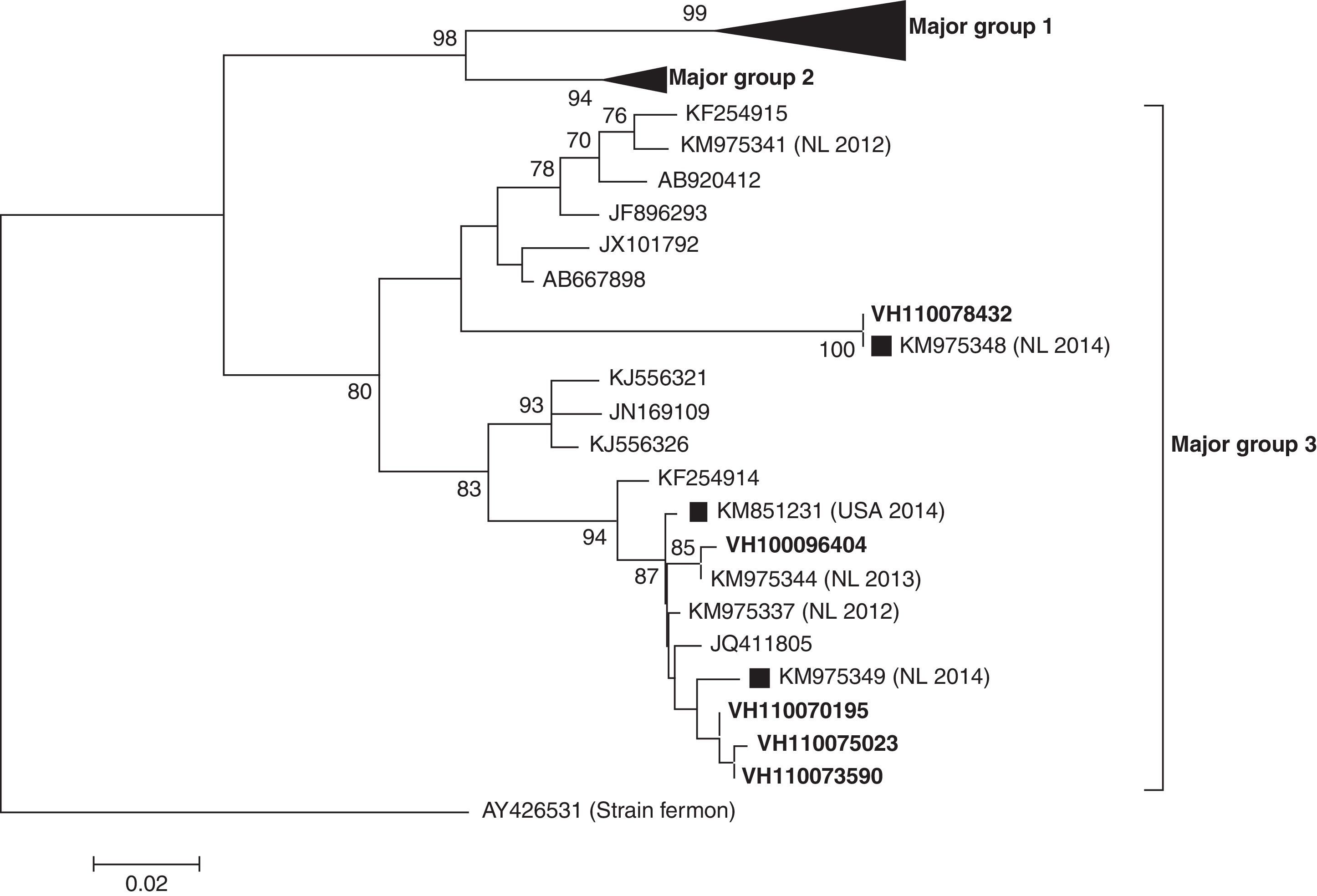
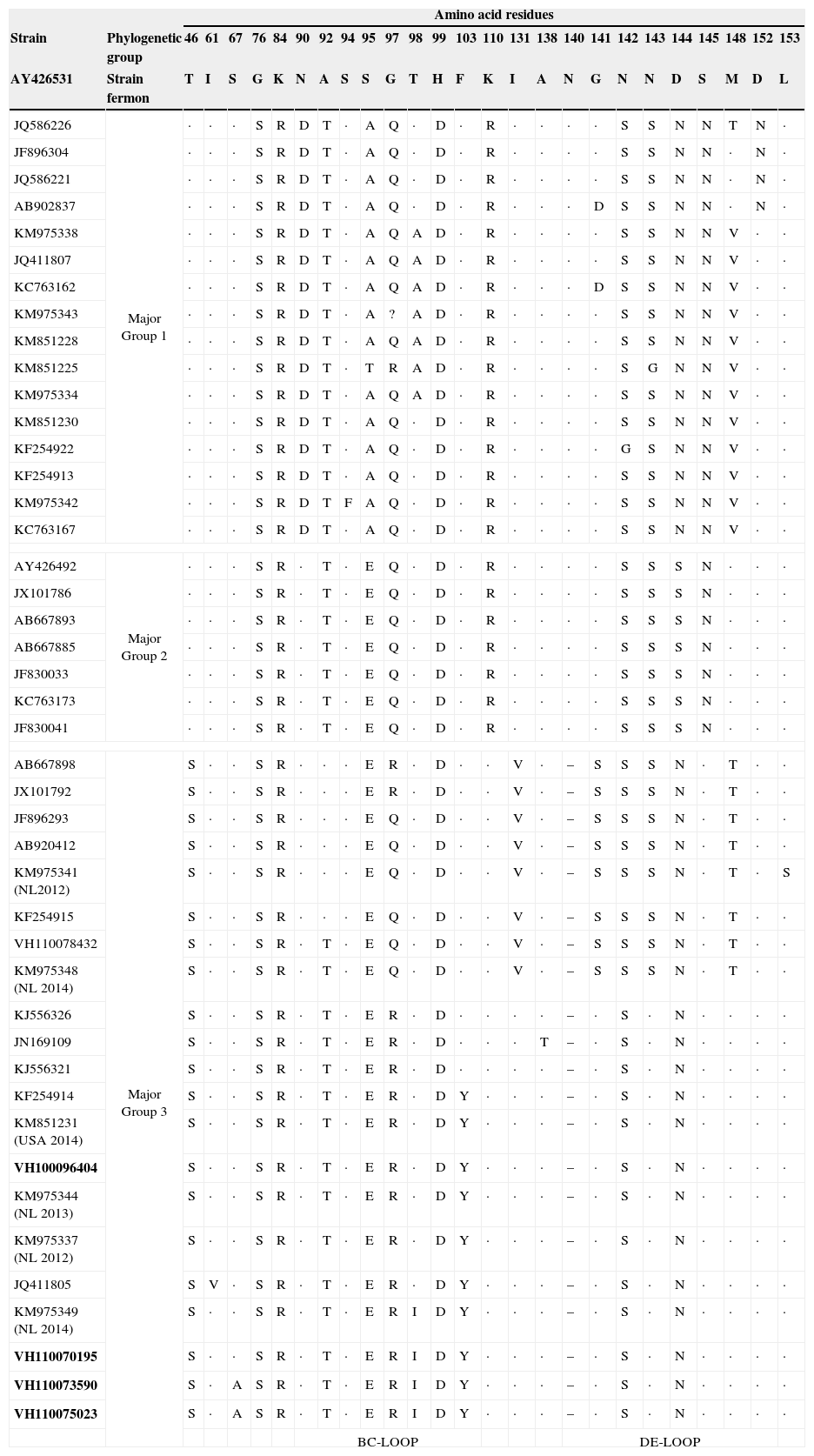
ResultsIn the period of study, a total of 209 samples from 163 patients were received for laboratory confirmation of respiratory viruses using in parallel both PCR-based methods. Samples from five patients were laboratory-confirmed for enterovirus using Cepheid SmartEV kit, but non-detected using Seegene Anyplex II RV16 Detection kit v1.1. In addition, samples from 50 patients were laboratory-confirmed for RV (38/50), hAdV (4/50), hCoV-229E (1/50), hBoV (2/50) and hPIV-4 (4/50). Phylogenetic analysis of partial VP1 sequences (Fig. 1) confirmed that all detected enteroviruses were D68 serotype, which belonged to the phylogenetic major group 3 among the three clades circulating worldwide, as previously described.3,4,8 The VP1 nucleotide sequences were genetically close to the recent 2014 Dutch strains4 and to the strains of the large 2014 USA outbreak. In the comparison of VP1 amino acid sequences of strains belonging to the different EV-D68 clades (Table 1), the EV-D68 strains that fell into the major group 3 show some amino acid substitutions, including an amino acid deletion, with most of the differences located in the immunogenic BC- and DE-loops.9 The nucleotide sequences of the present study were submitted to GenBank Database with accession numbers KP090456–KP090459 and KP122208.
Phylogenetic tree of partial (from nucleotide position 133–471) VP1 coding sequences of EV-D68 detected (in bold) in the present study. Sequences of recent Dutch (NL 2014) and USA (USA 2014) strains are marked with black squares. The evolutionary distances were computed using the Tamura 3-parameter method in MEGA v6, with a gamma distribution (shape parameter=0.49).
Amino acid substitutions of the deduced partial (from amino acid position 45 to 157) VP1 protein sequences from EV-D68 strains of the present study (in bold) relative to the homologous sequence of Fermon strain. Amino acids common to the Fermon strain are indicated with a dot in the alignment. Particular amino acid deletion (–) of sequences from strains belonging to major group 3 is shown by grey. The BC- and DE-loop regions in VP1 are boxed. Sequences of recent Dutch and USA strains are marked as NL 2014 and USA 2014, respectively.
| Amino acid residues | ||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Strain | Phylogenetic group | 46 | 61 | 67 | 76 | 84 | 90 | 92 | 94 | 95 | 97 | 98 | 99 | 103 | 110 | 131 | 138 | 140 | 141 | 142 | 143 | 144 | 145 | 148 | 152 | 153 |
| AY426531 | Strain fermon | T | I | S | G | K | N | A | S | S | G | T | H | F | K | I | A | N | G | N | N | D | S | M | D | L |
| JQ586226 | Major Group 1 | · | · | · | S | R | D | T | · | A | Q | · | D | · | R | · | · | · | · | S | S | N | N | T | N | · |
| JF896304 | · | · | · | S | R | D | T | · | A | Q | · | D | · | R | · | · | · | · | S | S | N | N | · | N | · | |
| JQ586221 | · | · | · | S | R | D | T | · | A | Q | · | D | · | R | · | · | · | · | S | S | N | N | · | N | · | |
| AB902837 | · | · | · | S | R | D | T | · | A | Q | · | D | · | R | · | · | · | D | S | S | N | N | · | N | · | |
| KM975338 | · | · | · | S | R | D | T | · | A | Q | A | D | · | R | · | · | · | · | S | S | N | N | V | · | · | |
| JQ411807 | · | · | · | S | R | D | T | · | A | Q | A | D | · | R | · | · | · | · | S | S | N | N | V | · | · | |
| KC763162 | · | · | · | S | R | D | T | · | A | Q | A | D | · | R | · | · | · | D | S | S | N | N | V | · | · | |
| KM975343 | · | · | · | S | R | D | T | · | A | ? | A | D | · | R | · | · | · | · | S | S | N | N | V | · | · | |
| KM851228 | · | · | · | S | R | D | T | · | A | Q | A | D | · | R | · | · | · | · | S | S | N | N | V | · | · | |
| KM851225 | · | · | · | S | R | D | T | · | T | R | A | D | · | R | · | · | · | · | S | G | N | N | V | · | · | |
| KM975334 | · | · | · | S | R | D | T | · | A | Q | A | D | · | R | · | · | · | · | S | S | N | N | V | · | · | |
| KM851230 | · | · | · | S | R | D | T | · | A | Q | · | D | · | R | · | · | · | · | S | S | N | N | V | · | · | |
| KF254922 | · | · | · | S | R | D | T | · | A | Q | · | D | · | R | · | · | · | · | G | S | N | N | V | · | · | |
| KF254913 | · | · | · | S | R | D | T | · | A | Q | · | D | · | R | · | · | · | · | S | S | N | N | V | · | · | |
| KM975342 | · | · | · | S | R | D | T | F | A | Q | · | D | · | R | · | · | · | · | S | S | N | N | V | · | · | |
| KC763167 | · | · | · | S | R | D | T | · | A | Q | · | D | · | R | · | · | · | · | S | S | N | N | V | · | · | |
| AY426492 | Major Group 2 | · | · | · | S | R | · | T | · | E | Q | · | D | · | R | · | · | · | · | S | S | S | N | · | · | · |
| JX101786 | · | · | · | S | R | · | T | · | E | Q | · | D | · | R | · | · | · | · | S | S | S | N | · | · | · | |
| AB667893 | · | · | · | S | R | · | T | · | E | Q | · | D | · | R | · | · | · | · | S | S | S | N | · | · | · | |
| AB667885 | · | · | · | S | R | · | T | · | E | Q | · | D | · | R | · | · | · | · | S | S | S | N | · | · | · | |
| JF830033 | · | · | · | S | R | · | T | · | E | Q | · | D | · | R | · | · | · | · | S | S | S | N | · | · | · | |
| KC763173 | · | · | · | S | R | · | T | · | E | Q | · | D | · | R | · | · | · | · | S | S | S | N | · | · | · | |
| JF830041 | · | · | · | S | R | · | T | · | E | Q | · | D | · | R | · | · | · | · | S | S | S | N | · | · | · | |
| AB667898 | Major Group 3 | S | · | · | S | R | · | · | · | E | R | · | D | · | · | V | · | – | S | S | S | N | · | T | · | · |
| JX101792 | S | · | · | S | R | · | · | · | E | R | · | D | · | · | V | · | – | S | S | S | N | · | T | · | · | |
| JF896293 | S | · | · | S | R | · | · | · | E | Q | · | D | · | · | V | · | – | S | S | S | N | · | T | · | · | |
| AB920412 | S | · | · | S | R | · | · | · | E | Q | · | D | · | · | V | · | – | S | S | S | N | · | T | · | · | |
| KM975341 (NL2012) | S | · | · | S | R | · | · | · | E | Q | · | D | · | · | V | · | – | S | S | S | N | · | T | · | S | |
| KF254915 | S | · | · | S | R | · | · | · | E | Q | · | D | · | · | V | · | – | S | S | S | N | · | T | · | · | |
| VH110078432 | S | · | · | S | R | · | T | · | E | Q | · | D | · | · | V | · | – | S | S | S | N | · | T | · | · | |
| KM975348 (NL 2014) | S | · | · | S | R | · | T | · | E | Q | · | D | · | · | V | · | – | S | S | S | N | · | T | · | · | |
| KJ556326 | S | · | · | S | R | · | T | · | E | R | · | D | · | · | · | · | – | · | S | · | N | · | · | · | · | |
| JN169109 | S | · | · | S | R | · | T | · | E | R | · | D | · | · | · | T | – | · | S | · | N | · | · | · | · | |
| KJ556321 | S | · | · | S | R | · | T | · | E | R | · | D | · | · | · | · | – | · | S | · | N | · | · | · | · | |
| KF254914 | S | · | · | S | R | · | T | · | E | R | · | D | Y | · | · | · | – | · | S | · | N | · | · | · | · | |
| KM851231 (USA 2014) | S | · | · | S | R | · | T | · | E | R | · | D | Y | · | · | · | – | · | S | · | N | · | · | · | · | |
| VH100096404 | S | · | · | S | R | · | T | · | E | R | · | D | Y | · | · | · | – | · | S | · | N | · | · | · | · | |
| KM975344 (NL 2013) | S | · | · | S | R | · | T | · | E | R | · | D | Y | · | · | · | – | · | S | · | N | · | · | · | · | |
| KM975337 (NL 2012) | S | · | · | S | R | · | T | · | E | R | · | D | Y | · | · | · | – | · | S | · | N | · | · | · | · | |
| JQ411805 | S | V | · | S | R | · | T | · | E | R | · | D | Y | · | · | · | – | · | S | · | N | · | · | · | · | |
| KM975349 (NL 2014) | S | · | · | S | R | · | T | · | E | R | I | D | Y | · | · | · | – | · | S | · | N | · | · | · | · | |
| VH110070195 | S | · | · | S | R | · | T | · | E | R | I | D | Y | · | · | · | – | · | S | · | N | · | · | · | · | |
| VH110073590 | S | · | A | S | R | · | T | · | E | R | I | D | Y | · | · | · | – | · | S | · | N | · | · | · | · | |
| VH110075023 | S | · | A | S | R | · | T | · | E | R | I | D | Y | · | · | · | – | · | S | · | N | · | · | · | · | |
| BC-LOOP | DE-LOOP | |||||||||||||||||||||||||
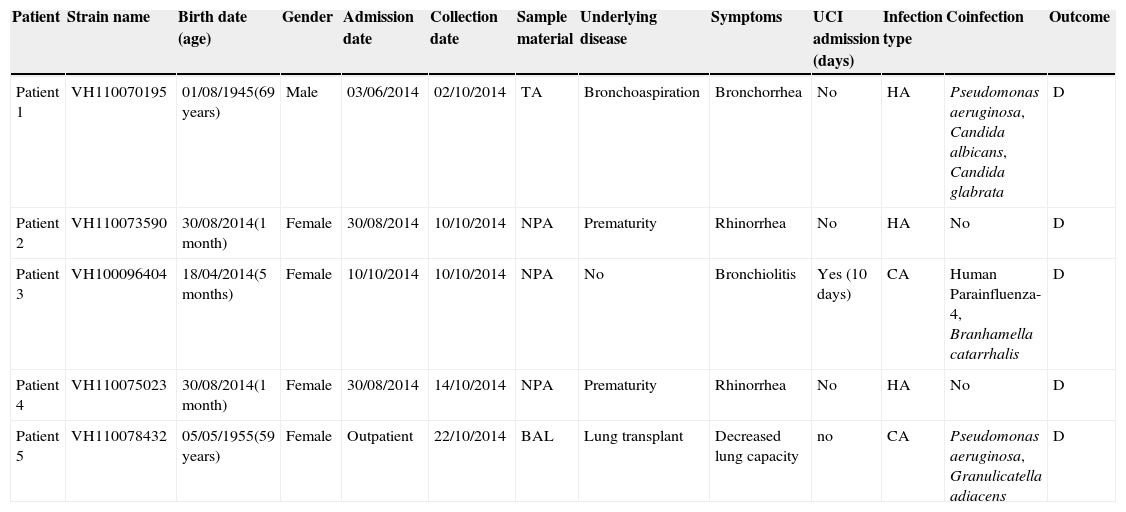
These EV-D68 strains were found in nasopharyngeal aspirates from three female infants less than one year of age, and in bronchoalveolar lavage and tracheal aspirate from two adults. Clinical and demographic features of EV-D68 infected patients are summarised in Table 2. All of them developed respiratory symptoms, but only one required Intensive Care Unit admission due to the severity of the infection. Coinfection was frequent, in three out of five patients. None of the patients described had symptoms of neurological disease or paralysis.
Summary of clinical and demographic data of the five EV-D68-confirmed cases of the present study.
| Patient | Strain name | Birth date (age) | Gender | Admission date | Collection date | Sample material | Underlying disease | Symptoms | UCI admission (days) | Infection type | Coinfection | Outcome |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient 1 | VH110070195 | 01/08/1945(69 years) | Male | 03/06/2014 | 02/10/2014 | TA | Bronchoaspiration | Bronchorrhea | No | HA | Pseudomonas aeruginosa, Candida albicans, Candida glabrata | D |
| Patient 2 | VH110073590 | 30/08/2014(1 month) | Female | 30/08/2014 | 10/10/2014 | NPA | Prematurity | Rhinorrhea | No | HA | No | D |
| Patient 3 | VH100096404 | 18/04/2014(5 months) | Female | 10/10/2014 | 10/10/2014 | NPA | No | Bronchiolitis | Yes (10 days) | CA | Human Parainfluenza-4, Branhamella catarrhalis | D |
| Patient 4 | VH110075023 | 30/08/2014(1 month) | Female | 30/08/2014 | 14/10/2014 | NPA | Prematurity | Rhinorrhea | No | HA | No | D |
| Patient 5 | VH110078432 | 05/05/1955(59 years) | Female | Outpatient | 22/10/2014 | BAL | Lung transplant | Decreased lung capacity | no | CA | Pseudomonas aeruginosa, Granulicatella adiacens | D |
Abbreviations: TA, tracheal aspirate; NPA, nasopharyngeal aspirate; BAL, bronchoalveolar lavage; HA, hospital-acquired; CA, community-acquired; D, discharged.
Since enteroviruses and rhinoviruses show a pattern of seasonality in the late summer and autumn in Northern Hemisphere, it is usual to detect these viruses in respiratory samples from hospitalised patients in September and October. In the majority of EV-D68 cases reported worldwide the children were the main population affected, but adults can also be infected, as seen in the present study.
The acute flaccid paralysis detected in the USA and Canada in children with EV-D68 infection has raised concerns about the aetiological role of this EV serotype in severe neurological disease.6 Recently, the first case of AFP following EV-D68 infection was reported in France.10 None of our EV-D68 infected patients had symptoms of neurological disease or paralysis, as also shown among patients of the last Dutch report.10 The severity of clinical symptoms seems to be related with the presence of underlying diseases. Therefore, four of the patients in our series had risk factors to develop severe respiratory infections, and EV-D68 acquisition was nosocomial in three patients, but only one suffered severe bronchiolitis that required mechanical ventilation (Table 2). Anyway, clinicians should be on the alert for severe respiratory illness in children that might be caused by infection with EV-D68 for an early diagnosis. Although there are no vaccines or specific treatments for EV-D68 infection, and clinical care is only supportive, the detection and identification of these viruses could help in explaining serious respiratory illness, giving guidance to medical care, preventing unnecessary treatment with antibiotics and adopting the isolation protocols to avoid the nosocomial transmission.
In the last 5 years the circulation of three different EV-D68 clades (major groups 1, 2 and 3) based on the phylogenetic analysis of VP1 sequence was reported.3,4,8 The EV-D68 strains characterised in this study belonged to the major group 3, and were similar to the strains recently described in The Netherlands that fell into this cluster.4 Some amino acid substitutions including an amino acid deletion at position 140 were found (Table 1) in the VP1 amino acid sequences from strains belonging to the major group 3 relative to Fermon strain, which differentiate them from the other strains belonging to the other two major groups. Since most of the amino acid differences fell into the BC- and DE-loops, these might be associated with changes in the antigenicity features, as previously described.9 However, an association between these virological features and the severity of infection cannot be established.
Finally, the genetic similarity between enterovirus and rhinovirus species makes the laboratory confirmation by molecular methods difficult due to the lack of specificity of primers and probes. Previously, in our laboratory Seegene's method was only used as the routine diagnostic method for the detection of respiratory viruses, including all EV serotypes, as commercially approved. But similar Seegene's products such as Seeplex® RV15ACE Detection kit (Seegene, Korea) did not seem to detect EV-D68 in a multi-centre study in the Netherlands.11 The decision to use it in parallel with a commercial EV-specific diagnostic test was made both to avoid false results as RV, and to assure the detection of the different known EV serotypes in order to accomplish with ECDC recommendations. The respiratory virus most frequently detected in the studied samples was RV, as expected for this period of the year, but none of these RV laboratory-confirmed samples were positive using Cepheid's EV-specific RT-PCR assay. It excluded the possibility that other EV, in particular EV-D68 cases, were wrongly identified as RV. However, the fact that Seegene's method is used by many European laboratories to detect EV in addition to other respiratory viruses suggests that the EV-D68 prevalence and its importance in Europe might therefore be underestimated.
In conclusion, additional evidence supporting the role of EV-D68 in respiratory infections in hospitalised Spanish patients has been provided. However, the fact that our institution is a tertiary referral hospital could have biased our findings towards patients with comorbidity. Therefore, enterovirus surveillance not only in hospitalised patients but also in outpatients through national networks for respiratory viruses surveillance should be enhanced in the following months.
Conflict of interestThe authors declare no conflicts of interest.
This work was partially supported by Plan Nacional de I+D+i 2008-2011 and Instituto de Salud Carlos III, Subdirección General de Redes y Centros de Investigación Cooperativa, Ministerio de Economía y Competitividad, Spanish Network for Research in Infectious Diseases (REIPI RD12/0015) – co-financed by European Development Regional Fund “A way to achieve Europe” ERDF.