Klebsiella pneumoniae carbapenemase (KPC)-producing bacteria are amongst the most important causative agents of nosocomial infections worldwide. Isolates of this bacterium have been identified in Venezuela but little is known about their local spread. The aim of this study was to perform the molecular characterization of KPC-producing strains isolated from 2012 to 2013 in a public hospital in Caracas, Venezuela.
MethodsTwenty-two K. pneumoniae clinical isolates phenotypically classified as KPC producing were subjected to PCR screening for the presence of blaKPC genes and their location within transposon Tn4401. The blaKPC PCR product was sequenced to identify the KPC alleles. Genotypic analysis was performed by means of repetitive extragenic palindromic PCR (rep-PCR) and Multi Locus Sequence Typing (MLST). Finally, conjugation and electroporation assays were used to determine whether the blaKPC genes were found in plasmids.
ResultsAll isolates contained the blaKPC-2 variant, and 21 of the 22 were associated with the Tn4401b isoform. The strains were distributed in 8 sequence types (ST), three of which were new. Conjugation and electroporation assays indicated that 95.5% (n=21/22) of the isolates contained the blaKPC gene in plasmids.
ConclusionsThis study on circulating bacterial strains and the identification of KPC alleles may help to understand the routes of dissemination and control their spread within this hospital.
Klebsiella pneumoniae productora de carbapenemasas tipo KPC es uno de los principales agentes causantes de infecciones nosocomiales a nivel mundial. En Venezuela se han identificado aislados de esta bacteria, sin embargo, se conoce poco sobre su dispersión. El objetivo de este estudio fue realizar la caracterización molecular de aislados de K. pneumoniae productores de KPC provenientes de un hospital público en Caracas, Venezuela, durante los años 2012 y 2013.
MétodosSe analizaron 22 aislados de K. pneumoniae clasificados fenotípicamente como productores de KPC, se les realizó la detección del gen blaKPC así como su ubicación en el transposón Tn4401 mediante PCR. El producto de PCR del gen blaKPC se secuenció para identificar los alelos circulantes. El análisis genotípico se realizó empleando las técnicas de amplificación por PCR de las secuencias repetidas extragénicas palindrómicas (rep-PCR) y la secuenciación de múltiples loci (MLST). Mediante ensayos de conjugación o electroporación, se determinó si los genes blaKPC se encontraban en plásmidos.
ResultadosLos 22 aislados contenían una variante del gen blaKPC-2 pero 21 de ellos estaban asociados a la isoforma Tn4401b y se distribuyeron en 8 secuencias tipo (ST), siendo tres de ellas nuevas. Los ensayos de conjugación y electroporación indicaron que el 95,5% (n=21/22) de los aislados portaban al gen blaKPC en plásmidos.
ConclusiónEl estudio de aislados clínicos circulantes así como la identificación de los alelos del gen blaKPC podría ayudar a conocer las rutas de diseminación y a controlar su propagación en este hospital.
Carbapenemase-producing Klebsiella pneumoniae (KPC) strains are increasingly reported worldwide.1 Bacteria producing these enzymes are generally susceptible only to a few remaining antibiotics and are associated with high mortality, especially in patients with bloodstream infections.2 In addition to being resistant to all β-lactams available, KPC carbapenemases have a high capacity to spread, as the KPC genes are contained within transposons such as Tn4401, which are carried on transferable plasmids.3,4 Although an alarming dispersion of carbapenem-resistant K. pneumoniae has been reported in the hospitals of several Venezuelan cities,5 the epidemiology and clinical impact of Venezuelan KPC-producing K. pneumoniae have not been described. The aim of this study was to characterize the predominant resistant clones circulating in a tertiary-care public hospital located in the Capital District of Caracas. Twenty two KPC-producing K. pneumoniae strains isolated from clinical specimens during the years 2012 and 2013 were studied to determine their evolutionary relatedness, identify their KPC alleles and the Tn4401 flanking regions and document the presence of blaKPC-carrying plasmids.
MethodsHospital settings“Dr. Domingo Luciani” is a large tertiary-care public hospital belonging to the Venezuelan Institute of Social Security that has 769 beds (727 inpatient and 42 intensive care), with a surgical preponderance. It is the principal public hospital in the eastern part of Caracas, the capital of Venezuela.
Study populationA cross-sectional study was conducted that included carbapenem-resistant K. pneumoniae strains isolated from January 2012 to December 2013. Molecular analyses were performed on the first clinical bacterial isolate recovered from a patient during their hospitalization. A total of 22 isolates were obtained, of which 16 were isolated in 2012 and 6 were isolated in 2013. Clinical and epidemiological information was obtained from the medical records for each patient and made irreversibly anonymous. This research had the approval of the Bioethics Committees of the Hospital and the Instituto Venezolano de Investigaciones Científicas.
Bacterial strains and antibiotic susceptibility testingThe study included twenty two K. pneumoniae isolates with intermediate or full resistant to carbapenems, according to CLSI 2012 cutoffs concentrations.6 The cut-off points for carbapenem sensitivity were Minimal Inhibitory Concentrations of ≤1μg/ml for meropenem and imipenem, and ≤0.5μg/ml for ertapenem. The species identification of isolates and determination of their antibiotic susceptibilities were performed with the automated MicroScan System (Siemens Healthcare Diagnostics Inc., CA, USA) in the hospital's clinical laboratory. All strains were tested for sensitivity to amikacin, gentamicin, imipenem, meropenem, ciprofloxacin, levofloxacin, tetracycline, tigecycline and trimethoprim/sulfamethoxazole. The modified Hodge Test was used to detect phenotypic carbapenemase production.7 For this test, the negative control was Escherichia coli ATCC 25922 and the positive control was a KPC – producing Klebsiella pneumoniae obtained from a previous study.8
Detection of genes encoding carbapenemasesPolymerase chain reaction (PCR) was performed with the primers listed in Table S1 to screen for the carbapenemase genes blaKPC,9blaIMP, blaVIM, blaNDM and blaOXA-48.10 After PCR amplification the fragments were sequenced with the corresponding primers in the forward and reverse directions by Macrogen, Korea, and the results were compared with sequences available in the Lahey database (www.lahey.org/Studies/) and GenBank®, using the Basic Local Alignment Search Tool (BLAST) (www.ncbi.nlm.gov/blast/).
Genetic environment of blaKPC geneThe genetic environment of the blaKPC gene was determined by PCR, using previously described primers11 specific for the Tn4401 transposon (Table S1).
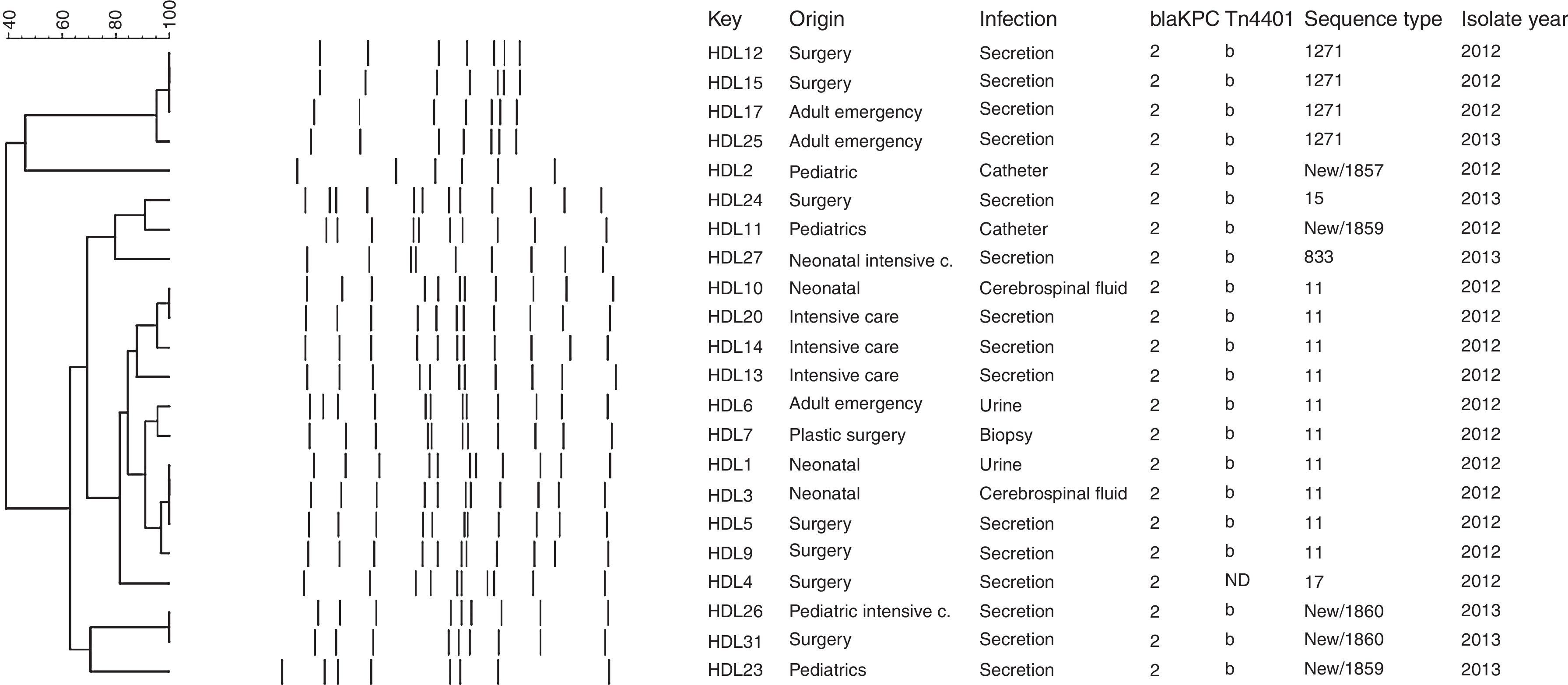
Molecular genotypingThe genetic relationships between the carbapenem-resistant isolates were determined by repetitive element palindromic-PCR (rep-PCR) and Multi-Locus Sequence Typing (MLST). rep-PCR was performed using previously described primers REP1 and REP2.12 PCR fingerprinting was analyzed with the BioNumerics software package version 7.5 (Applied Maths NV/Inc.) using the Dice binary coefficient and Unweighted Pair Group Method (UPGM) arithmetic average with settings of 1% tolerance and 1% optimization to create dendrograms. The positions of bands on each gel were normalized using the 100-bp DNA ladder (New England Biolabs® Inc.) as an external reference standard. MLST was performed as described by Diancourt et al.13 (Table S1). After PCR amplification, the fragments were sequenced with the corresponding primers in the forward and reverse directions by Macrogen, Korea. Allele numbers and sequence types (STs) were assigned by the Klebsiella pneumoniae MLST web site (http://www.pasteur.fr/recherche/genopole/PF8/mlst/Kpneumoniae.html).
Plasmid analysisConjugation experiments were performed to determine whether the carbapenemase resistance of the K. pneumoniae isolates was attributable to the carriage of blaKPC genes on plasmids. The recipient cells were Escherichia coli strain J-53 (F−, met−pro−, AziR). For the isolates from which transconjugants were not obtained, plasmid DNA was extracted by the Kieser extraction method14 and electroporated with a Bio-Rad Gene Pulser® into Escherichia coli XL1-Blue (F′, proAB, lacIqZΔM15, TcR) recipient cells. Transconjugants were selected on LB agar plates supplemented with ampicillin (100μg/ml) and sodium azide (200μg/ml), while transformants were selected on LB agar plates supplemented with ampicillin (100μg/ml). The presence of the blaKPC gene was confirmed by PCR.
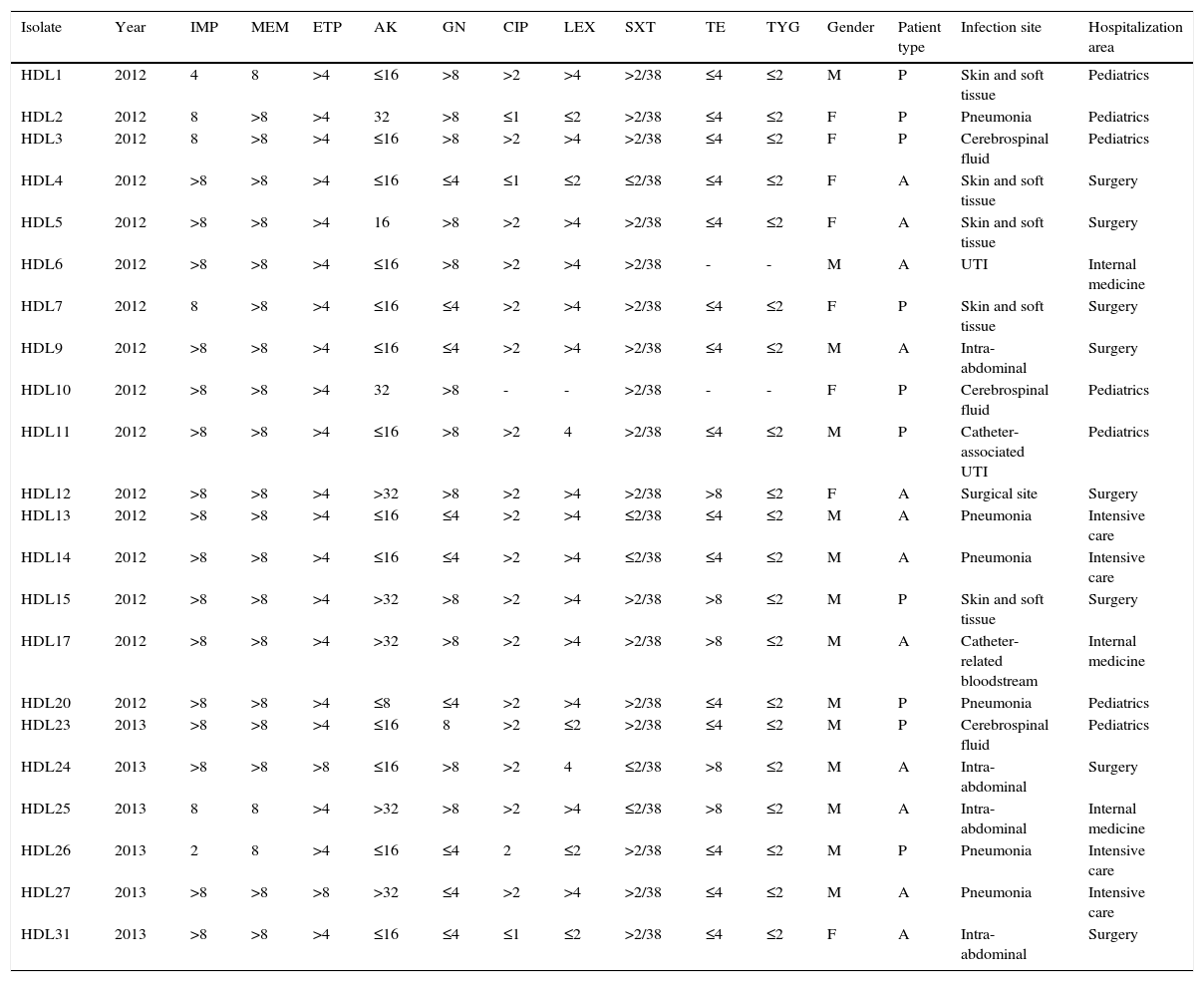
ResultsClinical and epidemiological characteristicsWe analyzed 22 carbapenem-resistant K. pneumoniae strains isolated from the clinical specimens of an equal number of inpatients in a public hospital in Caracas, Venezuela. The patients’ clinical characteristics are summarized in Table 1. Of the 22 total patients, 12 (54.5%) were adults and 14 (63.6%) were male. All infections were health care associated, with 8 (36.4%) from the surgical unit. The most common sites of infections were intra-abdominal, pneumonia, skin and soft tissue, with 5 patients each (22.7%), followed by catheter-related bloodstream and cerebrospinal fluid, with 2 patients each (9.2%) (Table 1).
Antibiotic susceptibility in KPC-producing K. pneumoniae isolates and clinical characteristics of patients from whom carbapenem-resistant K. pneumoniae isolates were obtained.
| Isolate | Year | IMP | MEM | ETP | AK | GN | CIP | LEX | SXT | TE | TYG | Gender | Patient type | Infection site | Hospitalization area |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HDL1 | 2012 | 4 | 8 | >4 | ≤16 | >8 | >2 | >4 | >2/38 | ≤4 | ≤2 | M | P | Skin and soft tissue | Pediatrics |
| HDL2 | 2012 | 8 | >8 | >4 | 32 | >8 | ≤1 | ≤2 | >2/38 | ≤4 | ≤2 | F | P | Pneumonia | Pediatrics |
| HDL3 | 2012 | 8 | >8 | >4 | ≤16 | >8 | >2 | >4 | >2/38 | ≤4 | ≤2 | F | P | Cerebrospinal fluid | Pediatrics |
| HDL4 | 2012 | >8 | >8 | >4 | ≤16 | ≤4 | ≤1 | ≤2 | ≤2/38 | ≤4 | ≤2 | F | A | Skin and soft tissue | Surgery |
| HDL5 | 2012 | >8 | >8 | >4 | 16 | >8 | >2 | >4 | >2/38 | ≤4 | ≤2 | F | A | Skin and soft tissue | Surgery |
| HDL6 | 2012 | >8 | >8 | >4 | ≤16 | >8 | >2 | >4 | >2/38 | - | - | M | A | UTI | Internal medicine |
| HDL7 | 2012 | 8 | >8 | >4 | ≤16 | ≤4 | >2 | >4 | >2/38 | ≤4 | ≤2 | F | P | Skin and soft tissue | Surgery |
| HDL9 | 2012 | >8 | >8 | >4 | ≤16 | ≤4 | >2 | >4 | >2/38 | ≤4 | ≤2 | M | A | Intra-abdominal | Surgery |
| HDL10 | 2012 | >8 | >8 | >4 | 32 | >8 | - | - | >2/38 | - | - | F | P | Cerebrospinal fluid | Pediatrics |
| HDL11 | 2012 | >8 | >8 | >4 | ≤16 | >8 | >2 | 4 | >2/38 | ≤4 | ≤2 | M | P | Catheter-associated UTI | Pediatrics |
| HDL12 | 2012 | >8 | >8 | >4 | >32 | >8 | >2 | >4 | >2/38 | >8 | ≤2 | F | A | Surgical site | Surgery |
| HDL13 | 2012 | >8 | >8 | >4 | ≤16 | ≤4 | >2 | >4 | ≤2/38 | ≤4 | ≤2 | M | A | Pneumonia | Intensive care |
| HDL14 | 2012 | >8 | >8 | >4 | ≤16 | ≤4 | >2 | >4 | ≤2/38 | ≤4 | ≤2 | M | A | Pneumonia | Intensive care |
| HDL15 | 2012 | >8 | >8 | >4 | >32 | >8 | >2 | >4 | >2/38 | >8 | ≤2 | M | P | Skin and soft tissue | Surgery |
| HDL17 | 2012 | >8 | >8 | >4 | >32 | >8 | >2 | >4 | >2/38 | >8 | ≤2 | M | A | Catheter-related bloodstream | Internal medicine |
| HDL20 | 2012 | >8 | >8 | >4 | ≤8 | ≤4 | >2 | >4 | >2/38 | ≤4 | ≤2 | M | P | Pneumonia | Pediatrics |
| HDL23 | 2013 | >8 | >8 | >4 | ≤16 | 8 | >2 | ≤2 | >2/38 | ≤4 | ≤2 | M | P | Cerebrospinal fluid | Pediatrics |
| HDL24 | 2013 | >8 | >8 | >8 | ≤16 | >8 | >2 | 4 | ≤2/38 | >8 | ≤2 | M | A | Intra-abdominal | Surgery |
| HDL25 | 2013 | 8 | 8 | >4 | >32 | >8 | >2 | >4 | ≤2/38 | >8 | ≤2 | M | A | Intra-abdominal | Internal medicine |
| HDL26 | 2013 | 2 | 8 | >4 | ≤16 | ≤4 | 2 | ≤2 | >2/38 | ≤4 | ≤2 | M | P | Pneumonia | Intensive care |
| HDL27 | 2013 | >8 | >8 | >8 | >32 | ≤4 | >2 | >4 | >2/38 | ≤4 | ≤2 | M | A | Pneumonia | Intensive care |
| HDL31 | 2013 | >8 | >8 | >4 | ≤16 | ≤4 | ≤1 | ≤2 | >2/38 | ≤4 | ≤2 | F | A | Intra-abdominal | Surgery |
IMP: imipenem, MEM: meropenem, ETP: ertapenem, AK: amikacin, GN: gentamicin, CIP: Ciprofloxacin, LEX: levofloxacin, SXT: trimethoprim-sulfamethoxazole, TE: tetracycline, TGC: tigecycline, (−): no data, (−): not detected, M: male, F: female, A: adult, P: pediatric, UTI, urinary tract infection.
All carbapenemase-producing K. pneumoniae isolates were resistant to ertapenem and meropenem while 21 (95.5%) were resistant to imipenem (Table 1). All isolates were positive with the Hodge Test, indicating the presence of a carbapenemase. The percentages of isolates resistant to the non-β-lactam drugs were as follows: gentamicin, 54.5% (n=12/22); amikacin, 22.7% (n=5/22); ciprofloxacin, 81% (n=17/21); levofloxacin, 66.6% (n=14/21); trimethoprim-sulfamethoxazole, 72.3% (n=17/22) and tetracycline, 20% (n=5/20), while 100% were susceptible to tigecycline (Table 1).
Detection of genes encoding carbapenemasesThe blaKPC-2 gene was detected in all isolates. None of the isolates harbored blaVIM, blaIMP, blaOXA-48 or blaNDM genes encoding others carbapenemases.
Genetic environment of blaKPC geneIn 21 of the 22 isolates (95.5%) from which blaKPC-2 gene was amplified, the Tn4401 primers produced the 703-bp PCR product associated with Tn4401 variant ‘b’.11 The ISKpn6 and tnpA genes were amplified and were also found in 21 of the 22 isolates. The inverted repeat sequences of the region flanking Tn4401 were not amplified with the specific primers described by Naas et al.4,15 There was only one isolate, HDL4, from which neither ISKpn6 nor the tnpA gene nor the nonconserved region of Tn4401 could be amplified.
Molecular genotyping of KPC-producing K. pneumoniae isolatesThe rep-PCR studies revealed that KPC-2-producing K. pneumoniae isolates belonged to several different genetic backgrounds (Fig. 1). MLST genotyping yielded 8 different sequence types: ST11 (45.5%, n=10), ST15 (4.5%; n=1), ST17 (4.5%; n=1), ST833 (4.5%; n=1), ST1271 (18.9%; n=4), and three novel sequence types that were deposited in the K. pneumoniae MLST database and assigned the designations ST1857 (4.5%; n=1), ST1859 (9.1%; n=2), and ST1860 (9.1%, n=2) (Fig. 1). The allelic profile of ST1857 is 17-19-39-20-125-4-305 and included a new tonB allele, while STs 1859 and 1860 have new alleles combinations (18-22-26-108-98-37-227 and 18-22-18-61-32-13-50, respectively), indicating that they are not SLV of a previously described Sequence Type.
Plasmid analysisNine of the 22 K. pneumoniae isolates (40.9%) transferred β-lactam resistance to E. coli recipients in the conjugation assay. Plasmid DNA was isolated from the remaining 13 K. pneumoniae isolates and used to electroporated the E. coli recipient strain, XL1-Blue. Transformants were obtained with plasmid DNA from 12 of the 13 (92.3%), confirming that the blaKPC genes were located on plasmids. Both electroporation and conjugation experiments were unsuccessful for one isolate (HDL11), despite repeated attempts. The presence of the blaKPC gene was confirmed by PCR in all transformants and transconjugants.
DiscussionCarbapenems have been the treatment of choice for infections caused by ESBL-producing Enterobacteriaceae, but an undesirable consequence has been the selection of carbapenem-resistant strains.16 Although carbapenem resistance in the Enterobacteriaceae is still unusual in Venezuela, KPCs are carried on mobile genetic elements and are easily disseminated, so the rapid detection of KPC-carrying bacteria is essential for effective therapeutic options and control measures.17 The present study integrated clinical and molecular data on K. pneumoniae strains isolated in one large public hospital in Venezuela, and determined the presence of different circulating genotypes and KPC alleles in order to help identify and control routes of dissemination. The majority of the 22 carbapenem-resistant K. pneumoniae strains were isolated from the pediatric and surgical services areas and they varied in their antibiotic sensitive or resistance profiles.
The blaKPC gene could be amplified from all isolates and although there have been twenty-three variants of the KPC enzyme reported (KPC-2 through KPC-24) (http://www.lahey.org/studies/other.asp#table1), sequencing of the blaKPC genes amplified from our isolates showed that all possessed the KPC-2 allele variant, one of the most extensively distributed worldwide,2,18 including in the South American countries of Colombia,19,20 Brazil21,22 and Argentina.23,24 There have been three previous reports of KPC-producing enterobacteria isolated in Venezuela. A KPC-2-producing Enterobacter cloacae isolated from tracheal secretions of a cancer patient in a private hospital in Caracas25 and a KPC-2-producing Klebsiella oxytoca found in a pediatric patient with pneumonia in the intensive care unit at Los Andes University Hospital, Mérida, Venezuela.17 Finally, in a cross sectional study, Marcano et al.5 analyzed enterobacteria from eight hospitals in Caracas isolated between October 15, 2009 and January 15, 2010, and found that 1.9% was K. pneumoniae with a KPC producing phenotype,5 but the alleles of KPC genes were not identified.
The blaKPC gene is generally associated with a Tn3-based transposon, Tn4401, composed of a transposase gene, a resolvase gene, the blaKPC gene and two insertion sequences (ISKpn6 and ISKpn7). Six isoforms (a–e) of Tn4401 have been described, which differ by polymorphisms located upstream of the blaKPC gene.26 We found the blaKPC-2 gene associated with Tn4401 variant “b” in all isolates tested except HDL4. The “b” isoform has been observed in the USA,18 Colombia,11 and Brazil.3 Similar to the report by Pereira et al.,3 the inverted repeat sequences of the flanking regions were not amplified in our isolates, suggesting that their insertion sites may be different from that of K. pneumoniae YC described by Naas et al.4 In the HDL4 isolate PCR did not amplify either ISKpn6, the tnpA gene, or the nonconserved region of Tn4401, suggesting that the genetic environment of the blaKPC-2 gene in this isolate is different from that of the other strains isolated from this hospital. A distinct genetic background of the blaKPC-2 gene has been identified in China, where it was found to be carried on plasmid pKP048. This plasmid contains the integration structure of a Tn3-based transposon and partial Tn4401 segment, with the gene order Tn3-transposase, Tn3-resolvase, ISKpn8, the blaKPC-2 gene and the ISKpn6-like element.27 Genotyping with rep-PCR showed that there were several K. pneumoniae genotypes circulating in the hospital, which was confirmed by MLST. The 22 blaKPC-2-harboring K. pneumoniae isolates belonged to 8 different sequence types, with 45% (n=10/22) belonging to ST11, part of the Clonal Complex CC258 that has been reported in many countries worldwide,11 including China,28 Korea,29 Hungary,30 Poland,31 Brazil,3,32 Spain,33 and United Kingdom.34 The second most common ST in our study, ST1271, found in 4 (18.9%) of our isolates, was previously reported in Cuba.35 This is particularly interesting because there are many Cuban medical personnel working in Venezuela. ST15 (4.5%; n=1), belonging to GC15, has been found in France, Italy, Spain and Israel,36 South Korea, Malaysia, Singapore and Thailand,37 Portugal,38 Hungary39 and China.40 ST833 (4.5%; n=1) has been reported in Israel36 and in Trieste, Italy41 and also is part of the Clonal Complex CC258. Interestingly, the KPC-2-producing Klebsiella pneumoniae ST833 strain was isolated in the Trieste Pediatric Hospital from the blood culture of a three year old patient transferred from a Venezuelan hospital to undergo marrow transplantation. In our on-going epidemiological study of KPC-producing K. pneumoniae in Venezuela, we have also found isolates belonging to ST833 in two other Venezuelan states: Zulia (in press) and Anzoátegui.8 ST17 has been found in China42 and Canada.43 It will be interesting to determine whether the novel sequence type we found – ST1857 (4.5%; n=1), ST1859 (9.1%; n=2), ST1860 (9.1%, n=2) – are widely distributed within Venezuela or merely locally restricted genotypes.
Plasmid analysis of the carbapenemase resistant K. pneumoniae isolates indicated that nine (40.9%) contained the blaKPC-2 gene on plasmids that were readily transferable by conjugation, while in twelve others a resistance conferring plasmid could only be transferred by electroporation. We were unable to transfer carbapenem resistance from strain HD11, with either conjugation or electroporation, although PCR and sequencing confirmed that it contained a blaKPC-2 gene. Perhaps the blaKPC gene in this strain is located on chromosome of this isolate, as described for Acinetobacter baumannii44 and Pseudomonas aeruginosa45 strains, but further studies are needed to confirm this possibility.
Antimicrobial resistance in Klebsiella pneumoniae is now a serious problem in many Venezuelan hospitals and requires urgent control measures, including strict adherence to hand hygiene and healthcare workers wearing gowns and gloves by when they enter rooms with patients having K. pneumonia-related illnesses. Healthcare facilities also must follow strict cleaning procedures to prevent the spread of K. pneumoniae. We hope that the molecular epidemiology provided by this study can aid to control the dissemination of the strains involved, but constant vigilance and on-going monitoring of resistant strains is essential for reducing their presence.
ConclusionsIn one large public hospital in Caracas, Venezuela, 22 carbapenem resistant K. pneumoniae containing the blaKPC-2 gene were isolated over a period of two years. These isolates were distributed into several different genotypes but 50% of them belong to CC258, most commonly as the widely distributed ST11, but three novel STs were also identified. This study underscores the need for collaborative institutional, local, national, and regional surveillance of KPC-producing K. pneumoniae to assist diagnosis and guide antimicrobial stewardship and infection control efforts.
FundingThis work was supported by research grants from Instituto Venezolano de Investigaciones Científicas (Number 1237), Caracas, Venezuela.
Conflict of interestThe authors declare that they have no conflicts of interest.
We thank the team of curators of the Institut Pasteur MLST and whole genome MLST databases for curating the data and making them publicly available at http://bigsdb.web.pasteur.fr/.