Central nervous system (CNS) infections such as meningitis and encephalitis are medical emergencies that require rapid diagnosis of the causative pathogen to guide early and adequate treatment since a delay in implementing an adequate antimicrobial therapy can lead to death. The current microbiological diagnostic methods based on culture or antigen detection have important limitations in their capacity to accurately identify the different potential pathogens causing CNS and, in the time, to obtaining results. Rapid syndromic molecular arrays have been developed. The main advantage of using a meningoencephalitis panel based in a multiplex test is that includes bacteria, viruses and fungi, covering the most prevalent microorganisms causing meningitis and encephalitis and the turn-around time is circa 1h. The use of these multiplex-PCR based tools is reviewed and the advantages and disadvantages of this technique are discussed.
Las infecciones del sistema nervioso central (SNC), tales como meningitis y encefalitis, son emergencias médicas que requieren un diagnóstico rápido del patógeno causante para orientar el tratamiento temprano y adecuado, ya que el retraso en la implementación de una terapia antimicrobiana adecuada conduce a una alta mortalidad. Los métodos actuales de diagnóstico microbiológico basados en el cultivo o la detección de antígenos tienen limitaciones importantes en su capacidad para identificar los diversos patógenos, en la precisión diagnóstica y en el tiempo hasta la obtención del resultado. Se han desarrollado ensayos rápidos sindrómicos basados en la PCR múltiple. La principal ventaja de usar un panel de meningoencefalitis es que incluye las bacterias, virus y hongos más prevalentes que causan meningitis y encefalitis y el tiempo de respuesta es de alrededor de 1hora. En este estudio se revisan estas herramientas basadas en la PCR múltiple, junto con las ventajas y desventajas que pueden mostrar.
In cases of suspected meningitis (inflammation of the meninges), encephalitis (inflammation of the cerebral parenchyma) or meningoencephalitis (inflammation of both the meninges and cerebral parenchyma) diagnosis of the aetiology of infection should be based on: 1. Personal epidemiological and clinical history: the predominant microorganisms are different in newborns and infants, children and adolescents, adults or in the elderly (Table 1); vaccinations received: Haemophilus influenzae b, Streptococcus pneumoniae, Neisseria meningitidis (C or A, C, Y, W135 and/or B), mumps (triple viral) and chickenpox; the presence of epidemic outbreaks; history of head trauma or neurosurgery; immunosuppression (HIV infection, transplantation, neoplasms, among others); history of travels to areas where certain infections are endemic; contact with animals or exposure to arthropod bites; coexistence of ear, nose or throat infections (otitis media, sinusitis or mastoiditis), mumps or sexually transmitted infections (syphilis or herpes simplex). 2. Symptomatology and laboratory data: the symptoms of these infections can be varied: fever, headache, meningism, altered consciousness, vomiting, seizures, neurological focus or rash, among others. The course of the disease (acute or subacute/chronic) will be different depending on the aetiology. Blood tests results can guide the diagnosis: leukocytosis with neutrophilia and elevation of C-reactive protein or procalcitonin are more frequent in bacterial meningitis. The presence of coagulation abnormalities can lead to meningococcal meningitis. Cytochemical analysis of cerebrospinal fluid (CSF) is essential: the white blood cell count (and morphology) and glucose and protein levels can suggest the presence of bacterial meningitis (a significant increase in leukocytes with a predominance of neutrophils, high proteins and low glucose) or viral meningitis (moderate increase in leukocytes with a predominance of lymphocytes and proteins, with normal glucose), or suspicion of a tuberculous or cryptococcal aetiology (moderate increase in leukocytes with a predominance of lymphocytes and proteins, with low glucose levels).
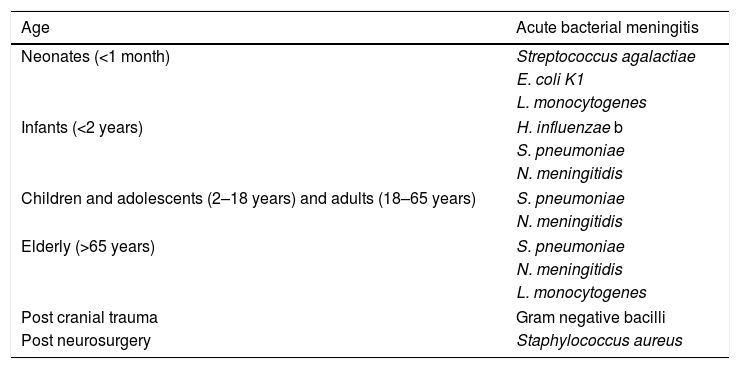
Aetiology of acute bacterial meningitis.
| Age | Acute bacterial meningitis |
|---|---|
| Neonates (<1 month) | Streptococcus agalactiae |
| E. coli K1 | |
| L. monocytogenes | |
| Infants (<2 years) | H. influenzae b |
| S. pneumoniae | |
| N. meningitidis | |
| Children and adolescents (2–18 years) and adults (18–65 years) | S. pneumoniae |
| N. meningitidis | |
| Elderly (>65 years) | S. pneumoniae |
| N. meningitidis | |
| L. monocytogenes | |
| Post cranial trauma | Gram negative bacilli |
| Post neurosurgery | Staphylococcus aureus |
Etiological diagnosis of meningitis and other central nervous system (CNS) infections could be achieve with different microbiological methods.
Conventional microbiological diagnosisThe Gram staining and culture of CSF are basic methods for the diagnosis of bacterial meningitis. In a Gram staining can show Gram negative coffee bean-shaped diplococci (N. meningitidis), Gram positive diplococci (S. pneumoniae), Gram negative cocobacilli (H. influenzae), Gram positive bacilli (Listeria monocytogenes), chain-sapped Gram positive cocci (Streptoccocus agalactiae) or Gram-negative bacilli (Escherichia coli). The observation of yeasts indicates meningitis due to Cryptococcus neoformans/gattii (capsulated yeasts in Chinese ink staining) or Candida spp. Therefore, the easy, rapid and inexpensive methodology of Gram staining is able to establish the aetiology of bacterial or fungal meningitis. The sensitivity of Gram staining of CSF ranges from 10 to 93% depending on the microorganism and bacterial load, and has a detection limit of approximately 104 colony forming units/ml.1 The use of enriched media (chocolate agar and blood agar) in CSF cultures can detect all these microorganisms in 24–48h. Blood cultures should be performed systematically on suspicion of bacterial meningitis or in the presence of high fever. Cultures using enriched media can also detect the growth of yeasts. Longer incubation times are needed for the less frequent meningitis due to filamentous, mainly dimorphic, fungi.
Ziehl-Neelsen or auramine stains are not very sensitive for detecting tuberculous meningitis, and therefore specific CSF culture for mycobacteria should be performed, although the sensitivity of this test is similarly low.2 CSF antigen detection techniques may be useful in patients with treated meningitis in which bacteria are observed in Gram staining and the CSF culture is negative. There are specific latex agglutination techniques for N. meningitidis (serotypes A, C, Y/W135), N. meningitidis serotype B/E. coli K1 (the two share antigen), S. pneumoniae, H. influenzae serotype b or S. agalactiae. Immunochromatography techniques to detect the S. pneumoniae antigen can also be used with the CSF.
The detection of the C. neoformans/gattii antigen by latex agglutination or immunochromatography, both in serum and in CSF, is very useful for the rapid diagnosis of meningitis by this microorganism.
CSF serological tests are helpful in the diagnosis of chronic neurological diseases due to syphilis (Treponema pallidum: VDRL) or Lyme disease (Borrelia burgdorferi: detection of IgG by enzyme-immunoassay or immunofluorescence and confirmation by Western blot).
Viral infections may also be due to different viruses as shown in Table 2. The diagnosis of viral infections is classically carried out by a combination of the CSF cell culture techniques and the detection of specific antibodies (IgG and IgM) in serum or CSF, however, these techniques have been surpassed by those of molecular biology which are described below.
The most frequent viral agents causing meningitis and encephalitis.
| Type of patient | Viruses |
|---|---|
| Immunocompetent | Enterovirus |
| Herpes simlex virus-1 and -2 | |
| Human Herpesvirus type-6 | |
| Measles | |
| Mumps | |
| Lymphocytic choriomeningitis virus | |
| Human Parechovirus | |
| Varicella-zoster | |
| Epstein–Barr virus | |
| Arboviruses | |
| Immunocompromised | HIV |
| JC virus | |
| Cytomegalovirus | |
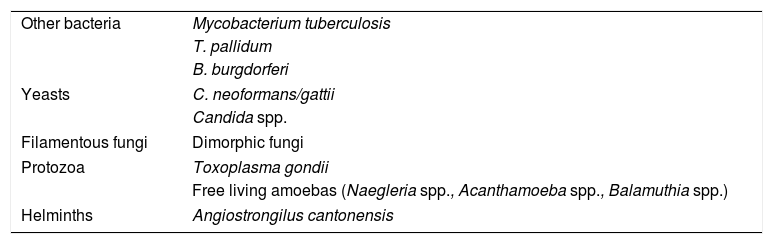
Meningoencephalitis is rarely caused by free-living amoebas such as Naegleria spp. (in individuals with a history of immersion in fresh water) and Acanthamoeba spp. or Balamuthia spp. (in immunosuppressed patients). In these cases, the diagnosis can be made by fresh examination or Giemsa stain of the CSF, or culture on an agar plate coated with bacteria (except for Balamuthia spp.). More rarely, meningoencepahilitis may be due to an infection by helminths such as Angiostrongilus cantonensis and occurs with eosinophilia (Table 3).
Other agents causing meningitis and encephalitis.
| Other bacteria | Mycobacterium tuberculosis |
| T. pallidum | |
| B. burgdorferi | |
| Yeasts | C. neoformans/gattii |
| Candida spp. | |
| Filamentous fungi | Dimorphic fungi |
| Protozoa | Toxoplasma gondii |
| Free living amoebas (Naegleria spp., Acanthamoeba spp., Balamuthia spp.) | |
| Helminths | Angiostrongilus cantonensis |
The drawbacks of the abovementioned tools have let to the development of methods of molecular diagnosis (nucleic acid amplification test – NAAT) mainly based on real-time PCR to detect either specific (singleplex) or multiple pathogens (multiplex).
New molecular diagnosisInvasive infectious diseases and particularly infections of the central nervous system (CNS) infections are medical emergencies that require rapid diagnosis of the causative pathogen in order to orient early and adequate treatment prevent life-long disabilities and save lives. Mortality by bacterial meningitis is higher than that viral meningitis3 and the delay in implementing an adequate antibiotic therapy leads to higher mortality.4,5
Current microbiological diagnostic methods based on culture or antigen detection have important limitations in their capacity to identify the diverse potential pathogens causing invasive infectious diseases, diagnostic accuracy, and time to achieving results. The utility of culture seems to be limited by the fact that the majority of potential human pathogens cannot be cultured, while those which can be cultured have a moderate sensitivity and the methodology is time-consuming. The main drawback of rapid antigen detection assays is that they are only suitable for testing pathogens that have previously been suspected of being a potential cause of the systemic disease and therefore, do not allow a blind and global detection approach. Overall, the aetiology of encephalitis6 and meningitis7 remains unknown in around 50% and 68% of cases in developed countries, respectively, leading to delayed or ineffective treatments, increased morbidity and mortality, suboptimal use of healthcare resources, and excessive diagnostic and treatment costs. Moreover, it should also be taken into account that by being live microorganisms pathogens are continuously adapting and evolving allowing them to elude the action of antimicrobials, the main therapeutic arsenal available to combat infections.
Singleplex assaySeveral singleplex assays for qualitative detection of different viruses such as enterovirus and herpes simplex virus (HSV) have been marketed. The advantages of the enterovirus detection kit from GeneXpert (Cepheid, Sunnyvale, Ca) over tissue cultures of viruses are that it has a sensitivity of 94–100%, it is a rapid test with a turnaround time of circa 2.5h and, in addition, it can detect viruses in 8–29% of the culture negative CSF.8,9 Overall, cell culture to isolate HSV from CSF has a poor sensitivity. The HSV-1 and -2 from CSF can be differentiated using a real-time PCR approach (DiaSorin Molecular, Cypress, CA). The turnaround time of this test is 1h and it has a sensitivity of 96–98% compared to the in-house developed real-time PCR.10,11 An important advantage of this test is that it only requires 50μl of CSF, and even with 20μl the sensitivity is still very high (96%). Although rare, mixed CSF infections caused either by two bacteria, two viruses and a bacterium and a virus have been described, thereby highlighting the potential risks of initial screening using singleplex PCR.10 In addition, another limitation of using singleplex assay is that the plethora of microorganisms causing meningitis and encephalitis make it difficult to rely on the symptoms to infer the causative agent of the infection.
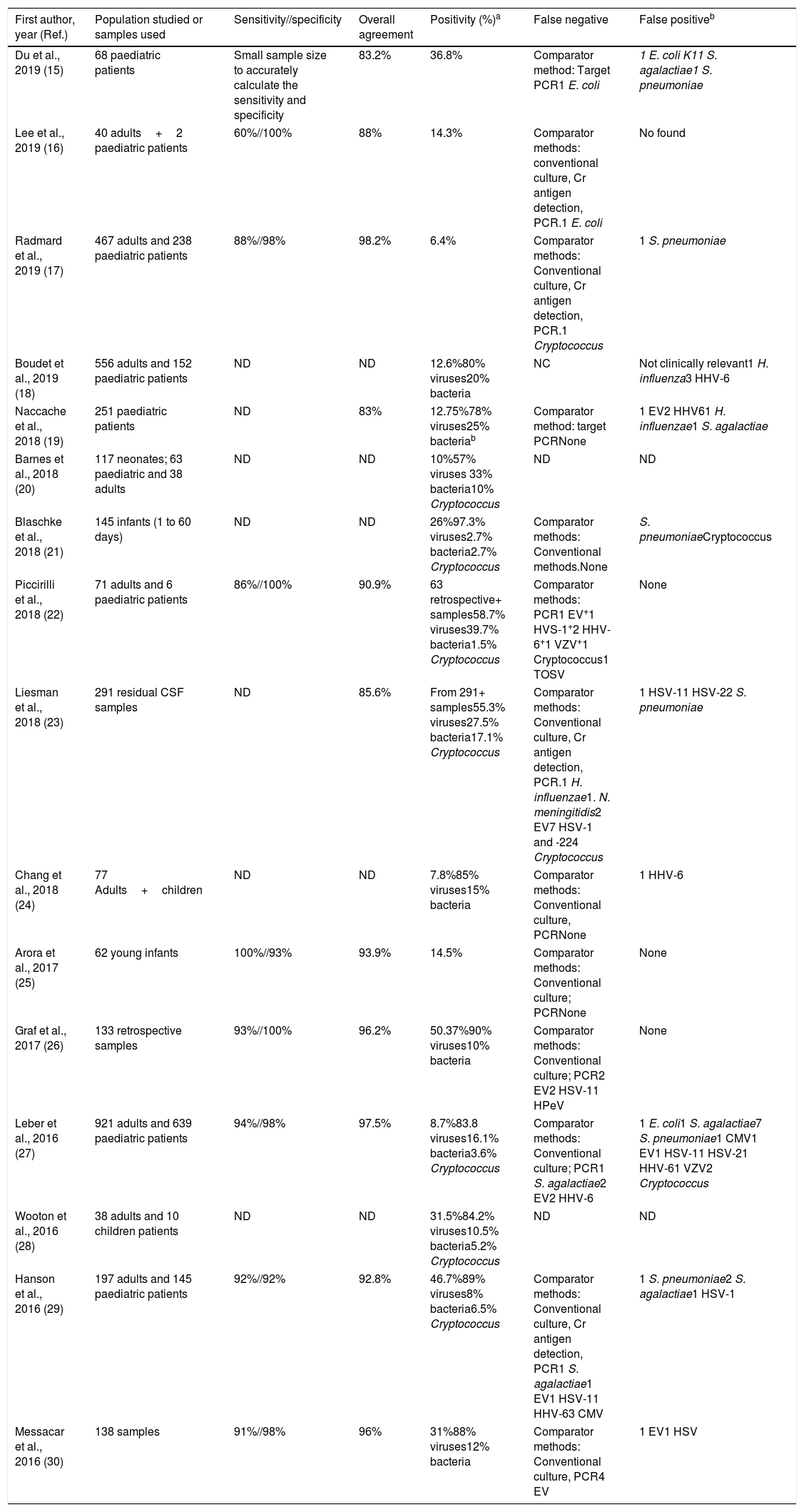
Multiplex testRapid syndromic molecular arrays covering the major known pathogens causing CNS infections have been developed. The main advantage of using a meningoencephalitis panel (ME panel) based in a multiplex test is that it includes both bacteria, viruses and fungi, covering the most prevalent microorganisms causing meningitis and encephalitis. The first commercialized and FDA approved panel to detect pathogens causing meningitis and encephalitis was Filmarray Meningitis/Encephalitis – ME (BioFire Diagnostics, Salt Lake City, UT). This panel covers 6 bacteria, 7 viruses and 1 fungi, including the following targets: E. coli K1, H. influenza, L. monocytogenes, N. meningitidis, Streptococcus agalactiae, S. pneumoniae, Cytomegalovirus, Enterovirus, Herpes simplex virus 1, Herpes simplex virus 2, Human herpes virus 6, Human parechovirus, Varicella zoster virus and C. neoformans/gattii. The assay is easy to prepare, it requires 200μl of CSF, only 2min of hands-on-time and the turnaround time is circa 1h. The design of the array is an automated nested Multiplex PCR system that has shown a higher sensitivity to detect low viral-loads in comparison with single step Real-time RT-PCR procedures.12 A recent meta-analysis reported that the estimated sensitivity and specificity of using Filmarray ME with a 95% of confidence intervals were 90% and 97%, respectively.13 However, most of the studies currently published (Table 4) using this panel only included a small number of positive samples for some pathogens, for instance, in the largest study to date, Leber et al.27 described a statistically significant sensitivity for only 9 out of the 14 pathogens detected. Some concerns have been raised about the specificity of this test, and different studies have reported potential false positive results due to specific DNA detection in CSF without clinical suspicion of the causal pathogen and associated with delay in appropriate clinical diagnosis and treatment.14
Molecular test (Filmarray) to detect pathogens causing meningitis and encephalitis using a multiplex approach.
| First author, year (Ref.) | Population studied or samples used | Sensitivity//specificity | Overall agreement | Positivity (%)a | False negative | False positiveb |
|---|---|---|---|---|---|---|
| Du et al., 2019 (15) | 68 paediatric patients | Small sample size to accurately calculate the sensitivity and specificity | 83.2% | 36.8% | Comparator method: Target PCR1 E. coli | 1 E. coli K11 S. agalactiae1 S. pneumoniae |
| Lee et al., 2019 (16) | 40 adults+2 paediatric patients | 60%//100% | 88% | 14.3% | Comparator methods: conventional culture, Cr antigen detection, PCR.1 E. coli | No found |
| Radmard et al., 2019 (17) | 467 adults and 238 paediatric patients | 88%//98% | 98.2% | 6.4% | Comparator methods: Conventional culture, Cr antigen detection, PCR.1 Cryptococcus | 1 S. pneumoniae |
| Boudet et al., 2019 (18) | 556 adults and 152 paediatric patients | ND | ND | 12.6%80% viruses20% bacteria | NC | Not clinically relevant1 H. influenza3 HHV-6 |
| Naccache et al., 2018 (19) | 251 paediatric patients | ND | 83% | 12.75%78% viruses25% bacteriab | Comparator method: target PCRNone | 1 EV2 HHV61 H. influenzae1 S. agalactiae |
| Barnes et al., 2018 (20) | 117 neonates; 63 paediatric and 38 adults | ND | ND | 10%57% viruses 33% bacteria10% Cryptococcus | ND | ND |
| Blaschke et al., 2018 (21) | 145 infants (1 to 60 days) | ND | ND | 26%97.3% viruses2.7% bacteria2.7% Cryptococcus | Comparator methods: Conventional methods.None | S. pneumoniaeCryptococcus |
| Piccirilli et al., 2018 (22) | 71 adults and 6 paediatric patients | 86%//100% | 90.9% | 63 retrospective+ samples58.7% viruses39.7% bacteria1.5% Cryptococcus | Comparator methods: PCR1 EV+1 HVS-1+2 HHV-6+1 VZV+1 Cryptococcus1 TOSV | None |
| Liesman et al., 2018 (23) | 291 residual CSF samples | ND | 85.6% | From 291+ samples55.3% viruses27.5% bacteria17.1% Cryptococcus | Comparator methods: Conventional culture, Cr antigen detection, PCR.1 H. influenzae1. N. meningitidis2 EV7 HSV-1 and -224 Cryptococcus | 1 HSV-11 HSV-22 S. pneumoniae |
| Chang et al., 2018 (24) | 77 Adults+children | ND | ND | 7.8%85% viruses15% bacteria | Comparator methods: Conventional culture, PCRNone | 1 HHV-6 |
| Arora et al., 2017 (25) | 62 young infants | 100%//93% | 93.9% | 14.5% | Comparator methods: Conventional culture; PCRNone | None |
| Graf et al., 2017 (26) | 133 retrospective samples | 93%//100% | 96.2% | 50.37%90% viruses10% bacteria | Comparator methods: Conventional culture; PCR2 EV2 HSV-11 HPeV | None |
| Leber et al., 2016 (27) | 921 adults and 639 paediatric patients | 94%//98% | 97.5% | 8.7%83.8 viruses16.1% bacteria3.6% Cryptococcus | Comparator methods: Conventional culture; PCR1 S. agalactiae2 EV2 HHV-6 | 1 E. coli1 S. agalactiae7 S. pneumoniae1 CMV1 EV1 HSV-11 HSV-21 HHV-61 VZV2 Cryptococcus |
| Wooton et al., 2016 (28) | 38 adults and 10 children patients | ND | ND | 31.5%84.2% viruses10.5% bacteria5.2% Cryptococcus | ND | ND |
| Hanson et al., 2016 (29) | 197 adults and 145 paediatric patients | 92%//92% | 92.8% | 46.7%89% viruses8% bacteria6.5% Cryptococcus | Comparator methods: Conventional culture, Cr antigen detection, PCR1 S. agalactiae1 EV1 HSV-11 HHV-63 CMV | 1 S. pneumoniae2 S. agalactiae1 HSV-1 |
| Messacar et al., 2016 (30) | 138 samples | 91%//98% | 96% | 31%88% viruses12% bacteria | Comparator methods: Conventional culture, PCR4 EV | 1 EV1 HSV |
In addition, false negative samples have also been described in almost most of the studies in which a discriminatory test has been used (Table 4) and these can be found in viruses, bacteria and cryptococcus. The main explanation for these false negatives could be due to suboptimal sensitivity of the PCR for certain targets, emergence of mutations in the regions where primers hybridize and also to a low pathogen load.
Others studies have shown a better yield of Filmarray compared to conventional tests in the diagnosis of ME.31–33 Other panels apart from Filmarray have been commercialized and have a CE marking [Allplex™ Meningitis (Seegene, KOR); MeningoFinder® 2SMART (Pathofinder, NL); Fast-track diagnosis, which has different panels (acquired by Siemens, DE); CertTest, which also has several panels (CerTest Biotec, ES)]. However, few studies using these panels are available in the scientific literature. One study using the Seeplex Meningitis ACE Detection kit (5 bacteria and 7 viruses), described a high detection.34 In a recent study by Säll et al.35 comparing Filmarray and MeningoFinder® 2SMART (PathoFinder, Maastricht, The Netherlands) the recovery of pathogens was higher with Filmarray, although when the limit of detection set up by the manufacturer of MeningoFinder was decreased, then the differences were not as great.
The main advantages of using the ME panels are: (i) they produce rapid results which help in the implementation of the optimal therapy and patient management, thereby decreasing unnecessary antibiotic use, and (ii) patients who have previously received antimicrobial therapy may show negative results using a conventional approach such as bacterial culture. On the other hand, the main limitations of ME panels are: (i) pathogens other than those included in the ME panel cannot been detected. This scenario can be found for instance in immunocompromised patients or post-neurosurgical CNS. Moreover, different CNS infections such as Japanese encephalitis, leptospirosis and neurosyphilis cannot be detected using the current ME panels and (ii). Both false-positive and false negative results may be shown (Table 4).36–38 False positive results are likely associated with contamination, whereas false-negative may be due to low quantity of the pathogen. Moreover, HHV-6 can be integrated into the human genome, mainly in peripheral lymphocyte cells which can be present in CSF, being the prevalence of HHV-6 circa 1% of the general population. Therefore, there is uncertainty about the pathogenic capacity of the detected microorganism39,40. In addition, the PCR does not distinguish between viable and unviable microorganisms.
Cost-efficiency analyses of ME panels are also lacking, and the high cost of these panels in all patients with suspected meningoencephalitis is likely not cost-effective. These panels would be more effective in immunocompromised patients or in children basing the MEs on criteria of differential white cells count in CSF. A recent study by Lumley and colleagues41 they found that the white cells count had a poor sensitivity in neonates (46%; <30 days) and in infants (31%; 30 days to 12 months) compared with children and adults that was 100%. However, in a recent study, Precit and colleagues42 showed that the use of the Filmarray ME panel based on pleocytosis or other abnormal parameters would have result in missed diagnostic, mainly in the detection of viruses in healthy and immunocompromised patients. Therefore, more studies are need to determine the most effective use of ME panels.
Rapid diagnosis in children: importance and current approachesMeningitis and encephalitis in children remain a major global health problem despite comprehensive prevention through vaccination and extensive use of antimicrobial therapies. According to the World Health Organization, meningitis and neonatal sepsis combined are the second infectious cause of death in children under five with more than 3,700,000 episodes and 355,770 deaths in 2016.43 In the last years improvements in global health and vaccination have reduced mortality by all causes of infectious diseases worldwide, however the decline in paediatric meningitis is much lower than that of other infectious diseases. Between 1990 and 2017 death by meningitis in children under 5 years of age reduced by only 53% while other causes of death of infectious diseases such as diarrhoea reduced by more than 70%.44–47
The vast array of invasive pathogens often produces indistinguishable clinical presentations that require the use of multiple microbiological tests to establish the classical etiological diagnosis and make adequate decisions for antimicrobial prescription. There are a number of important challenges to meet for the diagnosis and epidemiological surveillance of CNS infections in children and adults, including: (1) the emergence of microorganisms which are multi-resistant to antibiotics; (2) the identification of unknown pathogens that may cause or be involved in the development of these diseases; and (3) outbreaks caused by emergent vector–borne pathogens that spread into new geographical regions not previously affected. In addition, specific challenges in children are: (4) the differential diagnosis between an unknown congenital infection and a metabolic disease in the neonatal period and it should be highlighted that – (5) the first years of life constitute a fundamental stage for the physical and neuro-cognitive development along childhood, the maturation of the immune system and the development and stabilization of the human microbiota. On one hand, it is noteworthy that around 75% of brain growth occurs during the first 1000 days of life, and the brain during childhood has enormous metabolic capacity comprising 5–10% of total body mass, accounting for up to 50% of the body's basal metabolic energy of the body.48 Therefore, a potential infection affecting a child's brain could have devastating implications in comparison to its repercussion in an adult brain. On the other hand, the immune system is immature in early childhood and the development and stabilization of the human microbiota plays a crucial role in its maturation through antigenic stimulation. Since initiation response in neonates and young children is limited, the invasion of CSF by the first colonizing microorganisms can also have critically important effects compared to invasive infection during adulthood. These factors may explain the differential burden and consequences of CNS infections in particular age groups after the introduction of a new virus variant in a population. In 2016 an outbreak of brainstem encephalitis associated with a new variant of enterovirus-A71 in Catalonia, Spain, was reported. While this outbreak in children was associated with severe clinical manifestations and utilization of important healthcare resources, in adults the outbreak had very few consequences.49,50
The major pathogens causing meningitis in children change with age (Table 1). Group B streptococci, E. coli and, less frequently, L. monocytogenes, which are normal inhabitants of the vagina and bacteria that normally reside in the mother's digestive tract, and are acknowledged as the most common etiological agents in newborns. Beyond the neonatal period the most prevalent pathogens identified as the origin of meningitis are S. pneumoniae and N. meningitidis, while, in turn, the prevalence of H. influenzae type b, which was a primary cause of the disease in the past, has dramatically reduced its prevalence thanks to extensive vaccination. With respect to encephalitis, a high variety of different pathogens such as HHV-6, enterovirus or human paraechovirus are typical agents of encephalitis.
Table 4 shows the main publications reporting the use of the Filmarray ME panel in the diagnosis of meningitis and encephalitis in adults and children. Overall, the results in children are similar to those found in the adult population. A recent report stated that the application of the Filmarray ME panel can help guide the clinicians and reduce antibiotic treatment in children with suspected meningitis or encephalitis.51
Future perspectives in the molecular diagnosis of meningitis and encephalitisThe emergence and re-emergence of microorganisms multi-resistant to antibiotics or the replacement phenomenon against vaccine strategies are critical global health problems that can be mitigated by implementing rapid and accurate methods for pathogen diagnosis and characterization. There is a need for novel rapid precision tests able to detect any microorganism present in a specimen present in a simple and inexpensive manner. Such tests should be fast and capable of identifying pathogens in real time. They should also provide information about antimicrobial resistance agents and virulence factors to better orientate clinical decision making on the optimal treatment as well as provide more comprehensive information to public health agencies for epidemiological surveillance. A turnaround time of 8h should also be considered as a threshold limit for determining antimicrobial resistance in order to ensure that new tests enable the prescription of the best antimicrobial drug available at an early stage of disease and always before administering a second dose of the drug initially prescribed. This would notably improve patient management and antimicrobial stewardship. Moreover, novel tests should be competitive in cost compared with currently available microbiological techniques to facilitate widespread use in clinical settings.
Clinical metagenomics-based diagnosis with the use of Next Generation Sequencing (mNGS) for unknown pathogens are an alternative for the rapid detection of bacterial, viral, parasites and fungal pathogens, as well as for molecular characterization of antimicrobial resistance, virulence markers, and vaccine targets. mNGS is an untargeted technology that not only detects etiologic agents at species and even at a strain level but also allows the discovery of new or emergent microbial agents. This capacity may be useful for implementing One Health strategies since most emerging infectious diseases are zoonotic in origin. In addition, mNGS provides information on antimicrobial resistance and virulence factors, which are fundamental for the development of new vaccine targets and the prescription of the most adequate antimicrobial drugs. By combining unbiased sequencing, rapid data analysis, and access to comprehensive reference databases, mNGS can be applied for hypothesis-free, universal pathogen detection, being promising for improving etiological diagnosis, early treatment and epidemiological surveillance of invasive infections. Preliminary studies in different clinical contexts have demonstrated the potential of mNGS for unbiased pathogen detection in diverse infections, such as neuroleptospirosis52 and meningoencephalitis caused by Balamuthia mandrillaris.53 However, high costs, time consuming sequencing processes, and slow, sophisticated data analysis tools have long hindered routine application of mNGS in the clinical setting. To date, the limitations for the widespread clinical use of mNGS have been the high cost of the acquisition of sequencing devices and data storage infrastructures, intensive bench hand-labour, complex bioinformatics analysis and low sensitivity due to the interference of human DNA in the sample. However, rapid reductions in cost and increased throughput of NGS instruments, improved library preparation workflows, and continuous advances in speed and ease of use of data analysis tools are progressively removing these barriers and bringing NGS tests within the scope of clinical laboratories for routine diagnosis. In contrast, specimen preparation and data analysis steps are still highly complex and often overlooked, primarily due to the unique challenges posed by highly variable specimen complexity and quality, the broad genetic diversity of microorganisms, and imperfect and often highly incomplete reference databases. Clinical microbiologists are familiar with many of these challenges because they are similar to those involving with the use of NAATs.54 While mNGS is only available for the complex interpretation of results in international reference centres, rapid syndromic molecular arrays are a reality in the majority of clinical microbiology departments.
However, at present the major challenge in relation to this infection is the emergence of vector–borne diseases that easily spread across the world via international travel and the subsequent establishment of vectors and pathogens in populations not previously exposed. Re-emergence of endemic vector–borne diseases may occur due to climate-driven changes in their geographic range and ecology. Zika virus, Lyme disease, West Nile virus, and other vector–borne diseases have been identified as emerging non-enteric zoonoses and the majority of these agents are not detected by current commercial syndromic molecular point-of care assays.
Rapid molecular syndromic tests and mNGS technologies have the ability to detect multiple pathogens in a blinded and comprehensive manner but pose a new organizational challenge for the correct interpretation of results, particularly in the context of coinfections. An adequate strategy to face the complexities of unbiased detection approaches may be the establishment of multi-disciplinary partnerships between paediatricians, infectious diseases specialists and microbiologists in hospitals to jointly interpret outcomes from these novel technologies and determine their clinical value. Adaptation and evolution of the role of microbiologists will be crucial to ensure appropriate translation of their potential to the clinical practice. At present, the ease of use of rapid syndromic tests by minimally trained laboratory technicians has enabled their extensive deployment in clinical laboratories. In parallel, progressive improvements in the simplicity of mNGS pipelines anticipate their clinical application in the near future. While the evolution towards a new diagnostic paradigm driven by syndromic testing and mNGS will certainly facilitate operational sample processing tasks, it will also require a more in depth knowledge of the natural history of infection and disease, pathogen replication characteristics, virulence factors, pathogen ecology, big data analysis and other aspects needed for appropriate interpretation of massive results generated by present and upcoming innovative microbiological technologies.
The future role of clinical microbiology specialists as relevant actors in the diagnosis of CNS and other infectious diseases will rely highly on their versatility to adapt to the new diagnostic paradigm and contribute value to all these aspects in a multi-disciplinary work setting. In this sense the impact of rapid diagnostic platforms in Antimicrobial Stewardship Programmes can contribute to decreasing the antibiotic overutilization. Nevertheless, it has been found that 68 and 25% of patients started on empiric antibiotic therapy for a meningitis remained on antibiotics after 24 and 48h, respectively.17 In addition, the interpretation of positive results using ME panels requires additional clinical evaluation.
Conflict of interestsThe authors declare that they have no conflicts of interest.
This work was supported by the Instituto de Salud Carlos III, Subdirección General de Redes y Centros de Investigación Cooperativa, Ministerio de Economía y Competitividad, Spanish Network for Research in Infectious Diseases (REIPI RD16/0016/0010) and Spanish Network of Epidemiology and Public Health, CIBERESP. It was also co-financed by European Development Regional Fund “A way to achieve Europe”. This work was also supported by award 2017 SGR 0809 from the Agència de Gestió d’Ajuts Universitaris i de Recerca of the Generalitat de Catalunya.