Methicillin-resistant Staphylococcus aureus (MRSA) is considered a major cause of healthcare-associated (HA) and community-acquired (CA) infections. Considering non-β-lactam susceptibility as a potential marker for mecC-MRSA and CA-MRSA, the aim of this study was to determine the frequency and the associated genetic lineages of non-beta-lactam-antibiotic susceptible MRSA (NBLS-MRSA) strains in a multicenter study in Spain.
MethodsA collection of 45 NBLS-MRSA strains recovered in the period from January to June 2016 from 12 Spanish hospitals was analyzed. Molecular typing through spa-type characterization, agr group and multi-locus-sequence typing was performed. Methicillin-resistant genes (mecA and mecC) as well as immune evasion cluster (scn-chp-sak-sea-sep, considering scn gene as the marker of IEC system) and Panton-Valentine leukocidin (PVL) genes were determined with PCR/sequencing.
ResultsThe NBLS-MRSA phenotype was uncommon in the 12 hospitals analyzed (NBLS-MRSA/MRSA frequency: 0.3%-7.7%). All strains contained the mecA gene (and none contained mecC). Twenty-two different spa-types were detected among NBLS-MRSA strains, with spa-t008/agr-I the most prevalent (27%). The main clonal complexes were (CC/%): CC8/42.2%, CC5/33.3% and CC30/4.4%, with ST8 and ST5 as the main sequence types. The PVL toxin was present in 38% of strains (with spa-types t008, t024, t019, t044, t068, t318 and t3060). The IEC genes were detected in 78% of strains: IEC type-B (n=17), type-F (n=16), type-A (n=1) and type-E (n=1); 10 MRSA isolates were scn-negative.
ConclusionThe NBLS-MRSA phenotype is uncommon in the analyzed hospitals; although no mecC-positive strains were detected, it could be a good marker for MRSA PVL-positive isolates (38%), frequently associated with CA-MRSA infections.
Staphylococcus aureus resistente a meticilina (SARM) es una de las principales causas de infecciones tanto relacionadas con la asistencia sanitaria como asociadas a la comunidad (AC). Considerando la sensibilidad a antibióticos no-β-lactámicos como marcador potencial de SARM-mecC y SARM-AC, el objetivo de este estudio fue determinar la frecuencia y líneas genéticas de cepas SARM sensibles a antibióticos no-β-lactámicos (SARM-SNBL) en un estudio multicéntrico en España.
MétodosSe analizaron 45 cepas SARM-SNBL procedentes de 12 hospitales obtenidas durante enero-junio de 2016. El tipado molecular se realizó mediante caracterización del spa-tipo, grupo agr y multi-locus-sequence typing. Mediante PCR/secuenciación se determinaron los genes: de resistencia a meticilina (mecA y mecC), del sistema de evasión inmune humano (scn-chp-sak-sea-sep, usando scn como marcador del sistema IEC) y de la leucocidina de Panton-Valentine (LPV).
ResultadosEl fenotipo SARM-SNBL fue infrecuente en los 12 hospitales analizados (frecuencia SARM-SNBL/SARM: 0,3-7,7%). Todas las cepas fueron mecA-positivas (ninguna mecC). Se detectaron 22 spa-tipos diferentes, siendo el spa-t008/agr-I el prevalente (27%). Los principales complejos clonales fueron (CC/%): CC8/42,2%, CC5/33,3% y CC30/4,4%, destacando las secuencias tipo ST8 y ST5 como mayoritarias. El 38% de las cepas fue LPV-positiva (spa-tipos t008, t024, t019, t044, t068, t318 y t3060). El 78% de las cepas fue IEC-positivo: tipo-B (n=17), tipo-F (n=16), tipo-A (n=1) y tipo-E (n=1); 10 aislados fueron scn-negativos.
ConclusiónEl fenotipo SARM-SNBL es poco frecuente en los hospitales analizados; aunque no se detectaron cepas mecC-positivas, este fenotipo puede ser un buen marcador de aislados SARM LPV-positivos, frecuentemente asociados a infecciones por SARM-AC.
Staphylococcus aureus can colonize the skin and nose of humans and animals, but it can also be an important opportunistic pathogen associated with a wide spectrum of diseases. This microorganism can acquire several antimicrobial resistance mechanisms, being methicillin resistance one of the most relevant. Methicillin-resistant S. aureus (MRSA) are capable of survive in presence of β-lactam antibiotics due to the acquisition of staphylococcal cassette chromosome mec elements (SCCmec) carrying the mecA gene.1 In 2011, a new variant of the mecA gene, designated mecC or mecALGA251, which also confers methicillin resistance, was identified.2 One of the most significant characteristics of mecC-MRSA is its usual susceptibility to non-β-lactam antibiotics.3 The mecC gene was initially detected in human and bovine populations in Denmark and the UK,2 but since then, this mechanism has been detected in MRSA isolates of humans and animals in many European countries.3,4 In Spain, only a few cases of human infections have been reported,5–7 although it has been detected in wild and livestock animals, as well as in water.4,8–12 Additionally, other PBP2a-encoding gene named mecB, often found in a transposon mec complex (Tn6045) in Macrococcus caseolyticus, has also been found in S. aureus, but to date, only one case in humans has been reported in Germany.13
It is well known that MRSA is a major cause of healthcare-associated (HA-MRSA) infections, and also has a main role in community-acquired (CA-MRSA) infections. Since 1990s, MRSA infections in individuals without contact with health institutions have been reported, with USA300 strain as the CA-MRSA epidemic clone in the United States (ST8-IV clone).14 There are some differences between HA-MRSA and CA-MRSA isolates, as the profile of antibiotic resistance. CA-MRSA usually carry smaller SCCmec elements containing less antimicrobial resistance genes and more virulence factors. Consequently, CA-MRSA strains are more susceptible to antibiotics others than β-lactams; moreover, they frequently carry the lukF/lukS genes encoding the Panton-Valentine leucocidin (PVL), a two-component system with a toxin with cytolytic activity. Meanwhile, HA-MRSA clones are usually multidrug-resistant and unfrequently produce PVL toxin.15
Susceptibility for non-β-lactam antibiotics in MRSA isolates could be a marker for both mecC mechanism and CA-MRSA variant. For this reason, we focused this study on determining the frequency of non-β-lactam-susceptible MRSA (NBLS-MRSA) throughout the analysis of the genetic lineages, methicillin resistance mechanism and PVL gene detection in isolates recovered from 12 Spanish hospitals during a six-month period.
MethodsSelection of strainsMRSA isolates recovered from clinical and epidemiological samples during a six-month period (January-June 2016) were subjected to antimicrobial susceptibility testing. In addition to β-lactams, other 14 agents were tested (erythromycin, clindamycin, ciprofloxacin, levofloxacin, tetracycline, trimethoprim/sulfamethoxazole, vancomycin, teicoplanin, linezolid, daptomycin, fusidic acid, mupirocin, gentamicin, and tobramycin). All NBLS-MRSA isolates were included in this multicenter study, where 12 hospitals located in seven regions of Spain took part (full names of hospitals in Table 1). A final collection of 45 NBLS-MRSA isolates was obtained from the different institutions, and they were transferred to the University of La Rioja (Logroño, Spain) for further characterization. All strains were subcultured for 24h at 37°C in brain–heart infusion (BHI) agar and were stored frozen at −80°C. The total number of S. aureus and MRSA isolates of different patients recovered in the 12 hospitals in the six-month period was recorded for analysis.
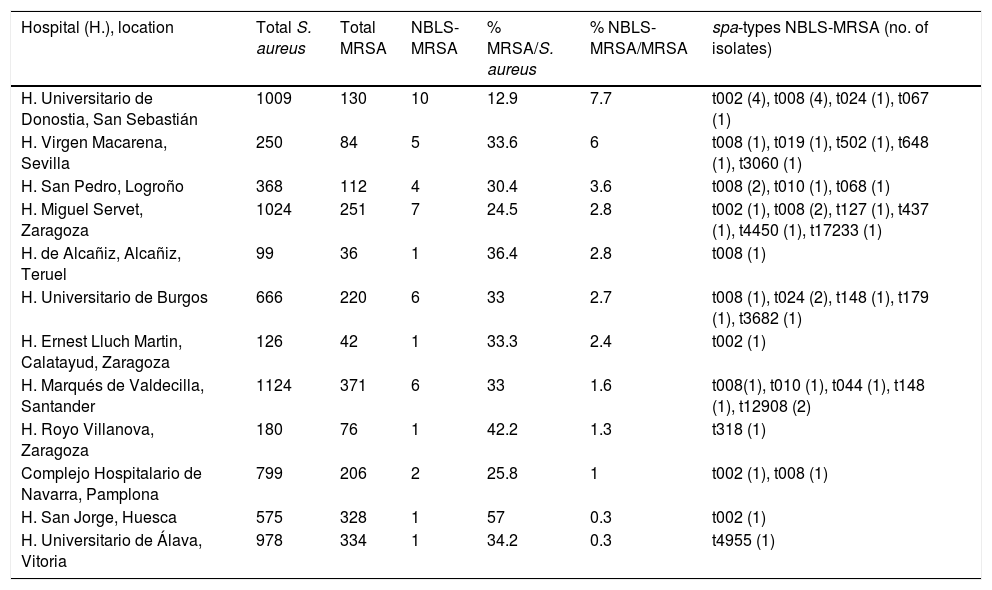
Distribution of isolates (number of S. aureus, MRSA and NBLS-MRSA), prevalence and NBLS-MRSA spa-types per hospital.
| Hospital (H.), location | Total S. aureus | Total MRSA | NBLS-MRSA | % MRSA/S. aureus | % NBLS-MRSA/MRSA | spa-types NBLS-MRSA (no. of isolates) |
|---|---|---|---|---|---|---|
| H. Universitario de Donostia, San Sebastián | 1009 | 130 | 10 | 12.9 | 7.7 | t002 (4), t008 (4), t024 (1), t067 (1) |
| H. Virgen Macarena, Sevilla | 250 | 84 | 5 | 33.6 | 6 | t008 (1), t019 (1), t502 (1), t648 (1), t3060 (1) |
| H. San Pedro, Logroño | 368 | 112 | 4 | 30.4 | 3.6 | t008 (2), t010 (1), t068 (1) |
| H. Miguel Servet, Zaragoza | 1024 | 251 | 7 | 24.5 | 2.8 | t002 (1), t008 (2), t127 (1), t437 (1), t4450 (1), t17233 (1) |
| H. de Alcañiz, Alcañiz, Teruel | 99 | 36 | 1 | 36.4 | 2.8 | t008 (1) |
| H. Universitario de Burgos | 666 | 220 | 6 | 33 | 2.7 | t008 (1), t024 (2), t148 (1), t179 (1), t3682 (1) |
| H. Ernest Lluch Martin, Calatayud, Zaragoza | 126 | 42 | 1 | 33.3 | 2.4 | t002 (1) |
| H. Marqués de Valdecilla, Santander | 1124 | 371 | 6 | 33 | 1.6 | t008(1), t010 (1), t044 (1), t148 (1), t12908 (2) |
| H. Royo Villanova, Zaragoza | 180 | 76 | 1 | 42.2 | 1.3 | t318 (1) |
| Complejo Hospitalario de Navarra, Pamplona | 799 | 206 | 2 | 25.8 | 1 | t002 (1), t008 (1) |
| H. San Jorge, Huesca | 575 | 328 | 1 | 57 | 0.3 | t002 (1) |
| H. Universitario de Álava, Vitoria | 978 | 334 | 1 | 34.2 | 0.3 | t4955 (1) |
The 45 NBLS-MRSA isolates were subjected to spa (S. aureus protein A) characterization by PCR16 and sequencing. The spa gene sequences were analyzed with Ridom® StaphType software17 (version 2.2.1). Determination of the accessory gene regulator (agr) group was performed by multiplex PCRs.18 The sequence type (ST) and clonal complex (CC) of selected isolates were determined by multilocus sequence typing (MLST),19 and for the other isolates the ST/CC was assigned according to their spa-types.
Detection of resistance genesThe presence of mecA and mecC methicillin-resistance genes as well as the penicillinase-encoding blaZ gene was studied by PCR.12
Detection of virulence factors (PVL and ACME) and the immune evasion cluster genesAll isolates were tested by PCR for the presence of lukF/lukS genes,20 encoding the PVL leucocidin. The two loci (arcA and opp3) that compose the arginine catabolic mobile element (ACME) was analyzed by PCR, as previously described,21 on the CC8 strains or other PVL-positive strains belonging to different clonal complexes. For the detection of the immune evasion cluster (IEC), the presence of five genes (scn, chp, sak, sea and sep) was analyzed.22 Attending to the combination of genes, the IEC could be ascribed to seven different groups (A-G).22
ResultsPrevalence of MRSA and NBLS-MRSA in the 12 studied hospitalsThe global rate of MRSA in the 12 hospitals included in the study in relation with S. aureus was of 30.4% (2190 MRSA out of 7198 S. aureus). Nevertheless, as it is shown in Table 1, important differences were found among hospitals (range: 12%–57%).
The phenotype NBLS-MRSA was very infrequent in the 12 hospitals included in the study (2.05% of total MRSA, and 0.63% of total S. aureus recovered in the six month-period), with differences among hospitals (range: 0.3%–7.7%) (Table 1).
Sample originOf the 45 NBLS-MRSA isolates included in this study, 80% were recovered from clinical samples and 20% of epidemiological surveillance (ES) samples. Within the group of clinical samples, 66.7% belonged to SSTI (skin and soft tissue infections) and 13.8% to urinary tract infections (UTI).
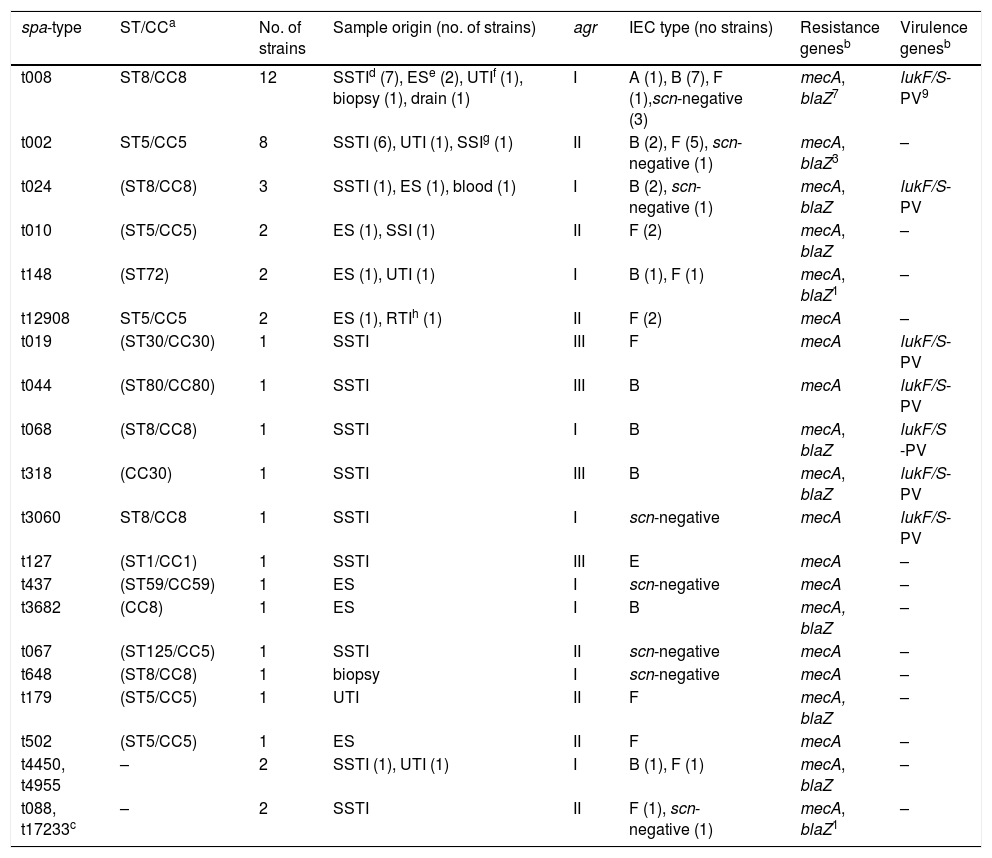
Molecular typing of MRSAThe spa-typing results of the 45 NBLS-MRSA strains are shown in Table 2, as well as its relation per hospital in Table 1. Twenty-two different spa-types were detected, being spa-t008/agr-I the most prevalent with 12 isolates (27%), as well as the most geographically extended (8 out of 12 centers). A new spa-type was identified (t17233) with the repeat succession 26–23–17–16-23–17–16. The main clonal complexes to which isolates were ascribed were the following ones: CC8 (42.2%), CC5 (33.3%), CC30 (4.4%), CC80 (2.2%), CC1 (2.2%) and CC59 (2.2%), with ST8 and ST5 as the predominant sequence types. Two isolates were ascribed to ST72 (spa-type t148). None of the strains was ascribed to CC130 or other clonal complexes usually related to the mecC gene.
Molecular typing, samples origin and genotypic characterization of the 45 NBLS-MRSA isolates.
| spa-type | ST/CCa | No. of strains | Sample origin (no. of strains) | agr | IEC type (no strains) | Resistance genesb | Virulence genesb |
|---|---|---|---|---|---|---|---|
| t008 | ST8/CC8 | 12 | SSTId (7), ESe (2), UTIf (1), biopsy (1), drain (1) | I | A (1), B (7), F (1),scn-negative (3) | mecA, blaZ7 | lukF/S-PV9 |
| t002 | ST5/CC5 | 8 | SSTI (6), UTI (1), SSIg (1) | II | B (2), F (5), scn-negative (1) | mecA, blaZ3 | – |
| t024 | (ST8/CC8) | 3 | SSTI (1), ES (1), blood (1) | I | B (2), scn-negative (1) | mecA, blaZ | lukF/S-PV |
| t010 | (ST5/CC5) | 2 | ES (1), SSI (1) | II | F (2) | mecA, blaZ | – |
| t148 | (ST72) | 2 | ES (1), UTI (1) | I | B (1), F (1) | mecA, blaZ1 | – |
| t12908 | ST5/CC5 | 2 | ES (1), RTIh (1) | II | F (2) | mecA | – |
| t019 | (ST30/CC30) | 1 | SSTI | III | F | mecA | lukF/S-PV |
| t044 | (ST80/CC80) | 1 | SSTI | III | B | mecA | lukF/S-PV |
| t068 | (ST8/CC8) | 1 | SSTI | I | B | mecA, blaZ | lukF/S -PV |
| t318 | (CC30) | 1 | SSTI | III | B | mecA, blaZ | lukF/S-PV |
| t3060 | ST8/CC8 | 1 | SSTI | I | scn-negative | mecA | lukF/S-PV |
| t127 | (ST1/CC1) | 1 | SSTI | III | E | mecA | – |
| t437 | (ST59/CC59) | 1 | ES | I | scn-negative | mecA | – |
| t3682 | (CC8) | 1 | ES | I | B | mecA, blaZ | – |
| t067 | (ST125/CC5) | 1 | SSTI | II | scn-negative | mecA | – |
| t648 | (ST8/CC8) | 1 | biopsy | I | scn-negative | mecA | – |
| t179 | (ST5/CC5) | 1 | UTI | II | F | mecA, blaZ | – |
| t502 | (ST5/CC5) | 1 | ES | II | F | mecA | – |
| t4450, t4955 | – | 2 | SSTI (1), UTI (1) | I | B (1), F (1) | mecA, blaZ | – |
| t088, t17233c | – | 2 | SSTI | II | F (1), scn-negative (1) | mecA, blaZ1 | – |
All the 45 NBLS-MRSA strains harbored the mecA gene and were mecC-negative (Table 2). The penicillinase encoded by the blaZ gene was present in 51% of strains (23/45). The IEC system was identified in 78% of the studied strains, and the remaining 10 strains lacked the scn gene and, in consequence, were considered as IEC negative (Table 2). Seventeen isolates carried the genes of IEC type-B (scn, chp, sak), 16 were IEC type-F positive (scn, chp, sak, sep), one strain IEC type-A (scn, chp, sak, sea) and another one type-E (scn, sak). Regarding the PVL virulence factor, 38% of the strains carried the lukF/S-PV genes, corresponding to the following spa-types: t008, t024, t068, and t3060 (all CC8), t044 (CC80), t019 and t318 (both CC30). The PVL encoding-genes were not detected in the isolates with the remaining spa-types identified in the study (Table 2). All CC8 or other PVL-positive strains lacked the ACME locus, typical of the USA300 clone.
DiscussionAccording to this study, MRSA with mecC genotype seems to be very infrequent in the analyzed hospitals, at least when the NBLS-MRSA marker was used for mecC-MRSA detection. In fact, all NBLS-MRSA strains carried the mecA gene. There are not many studies reflecting the presence of the mecC mechanism in MRSA human infections in Spain,5–7 and all of them report individual cases. The real prevalence of mecC is unknown in Europe, although in some countries mecC-MRSA isolates have increased (for instance, in Denmark from 1.8% in 2010 to 2.9% in 2011).23 A meta-analysis study on the prevalence of mecC-MRSA based on previously published results obtained until April 2015,24 suggested an estimated global mecC prevalence of 0.004% in humans and 0.1% in animals. In this way, our results confirm this low prevalence at hospital level. Nevertheless, we cannot discard the existence of mecC strains with resistance to non-β-lactam antimicrobials, very unusual at the present moment, but that has been occasionally described (for instance: two isolates of human origin resistant to ciprofloxacin,21 and one isolate recovered from wastewater resistant to erythromycin10). Moreover, mecC strains sometimes show borderline susceptibility results for oxacillin or cefoxitin, and could appear phenotypically as MSSA (methicillin-susceptible S. aureus).25 So, future studies could be focused in determining the presence of the mecC gene in S. aureus, with independence of the antimicrobial resistance phenotype (including β-lactams).
The zoonotic origin of mecC-MRSA is hypothesized since its origin in 2011 in cattle. Although the detection of mecC in humans is unusual, it presents a wide distribution in all animal species (livestock, companion or wildlife animals).3,4,8–11,26 Contact with animals might be a zoonotic risk,12 as mecC-MRSA can be easily transmitted between species.27
The NBLS-MRSA phenotype seems to be a good marker for PVL detection in MRSA isolates, considering that more than 1/3 of these strains were PVL-positive, while this factor is infrequent among non-selected MRSA isolates. The detected association of PVL production with the spa-type t008 (ST8/CC8), a classical CA-MRSA lineage28 and the most disseminated NBLS-MRSA in our country,29 is of relevance. In a previous study carried out in Spain, CA-MRSA corresponded to 2.9% of all studied MRSA obtained during the period 2004–2012 in the Spanish National Reference Centre of Staphylococci, and most of them showed susceptibility for non-β-lactams (84.5%), being most of them PVL-producers (91.9%).29
Observing the clonal complexes detected among NBLS-MRSA isolates, 42.2% belonged to CC8, which is strongly associated to the CA-MRSA USA300 clone,14,28 and 74% of them were PVL producers. On the other hand, none of the tested strains contained the ACME island. The other major clonal complex was CC5 (33.3%), a typical HA-MRSA, and none of these strains carried the genes for the PVL toxin, as expected. CC30 and CC80, both well-known CA-MRSA lineages,14,28 were present with 100% of PVL-positive strains. Two isolates with the spa-type t148, belonging to the CA-MRSA ST72, were detected. This clone is the most prevalent CA-MRSA in Korea causing infections, it was spread into hospital settings and it is also present in pigs and cattle carcasses.30 ST72 is not so frequent in Europe, although it has been detected more and more often in Spanish hospitals.21,29 Overall, at least 50% of CCs recorded in our study are associated to CA-MRSA, and the vast majority of strains with CCs related to CA-MRSA were PVL-producers.
Another important point is that NBLS-MRSA phenotype is very infrequent in the hospitals tested (0.3%–7.7%, media 2.05%). Therefore, it could be important to test the presence of the PVL genes when this phenotype is detected, mostly if strains are recovered from SSTI infections, due to the clinical relevance of this toxin. In our study, more than 75% of the NBLS-MRSA isolates that harbored the lukF/S-PV genes were isolated from SSTI.
The presence of the scn gene (marker of IEC system) in most NBLS-MRSA strains is expected, since its frequent detection among human isolates.31 Nevertheless, 22% of the studied strains were scn-negative, common feature of animal isolates. These scn-negative isolates belonged to many different STs and spa-types. Moreover, no relation between the production of PVL and the presence of the IEC system was observed: 11 out of the 17 PVL-producer strains had IEC type-B, two IEC type-F and four lacked the IEC genes. In the future it would be important to have epidemiological data of patients carrying scn-negative MRSA isolates to analyze the variables that could be associated to their acquisition.
Altogether, we can conclude that mecC-MRSA is very uncommon in human infections in the reported hospitals, but NBLS phenotype can be a valuable marker for PVL-producer strains (usually related to CA-MRSA), especially for the t008/agrI clone. It is important to maintain an active surveillance for these clones, not only for epidemiological control, but also for the right and early treatment in virulent PVL-infections.
Conflict of interestsThe authors declare that they have no conflicts of interest.
This work was supported by the Sociedad de Enfermedades Infecciosas del Norte (SEINORTE) and by the Agencia Estatal de Investigación (AEI) of Spain (project SAF2016-76571-R) and the Fondo Europeo de Desarrollo Regional (FEDER). Sara Ceballos and Laura Ruiz-Ripa have a predoctoral fellowship of the University of La Rioja, Spain.
Servicio de Microbiología, Hospital de Alcañiz, Alcañiz, Teruel (Jorge Arribas, Carmen Navarro);
Hospital Ernest Lluch Martin, Calatayud, Zaragoza (Antonina Arias, Blanca Fortuño);
Servicio Microbiología, Hospital Royo-Villanova, Zaragoza (Javier Pereira);
Servicio Microbiología, Hospital San Jorge, Huesca (Ana Milagro, Luis Torres);
Laboratorio Microbiología, Hospital San Pedro, Logroño (Carla Andrea Alonso, Luis Miguel Soria-Blanco);
Servicio de Microbiología, Hospital Universitario de Álava, Vitoria (Andrés Canut, M. Luz Cordón);
Servicio de Microbiología, Hospital Universitario de Burgos, Burgos (Gregoria Megías);
Servicio de Microbiología, Hospital Universitario Marqués de Valdecilla, Santander (Jorge Calvo);
Servicio de Microbiología, Hospital Universitario Miguel Servet/IIS Aragón, Zaragoza (Antonio Rezusta).