Enterococcus faecium has emerged as a multidrug-resistant nosocomial pathogen involved in outbreaks worldwide. Our aim was to determine the antimicrobial susceptibility, biofilm production, and clonal relatedness of vancomycin-resistant E. faecium (VREF) clinical isolates from two hospitals in Mexico.
MethodsConsecutive clinical isolates (n=56) were collected in two tertiary care hospitals in Mexico from 2011 to 2014. VREF isolates were characterized by phenotypic and molecular methods including pulsed-field gel electrophoresis (PFGE).
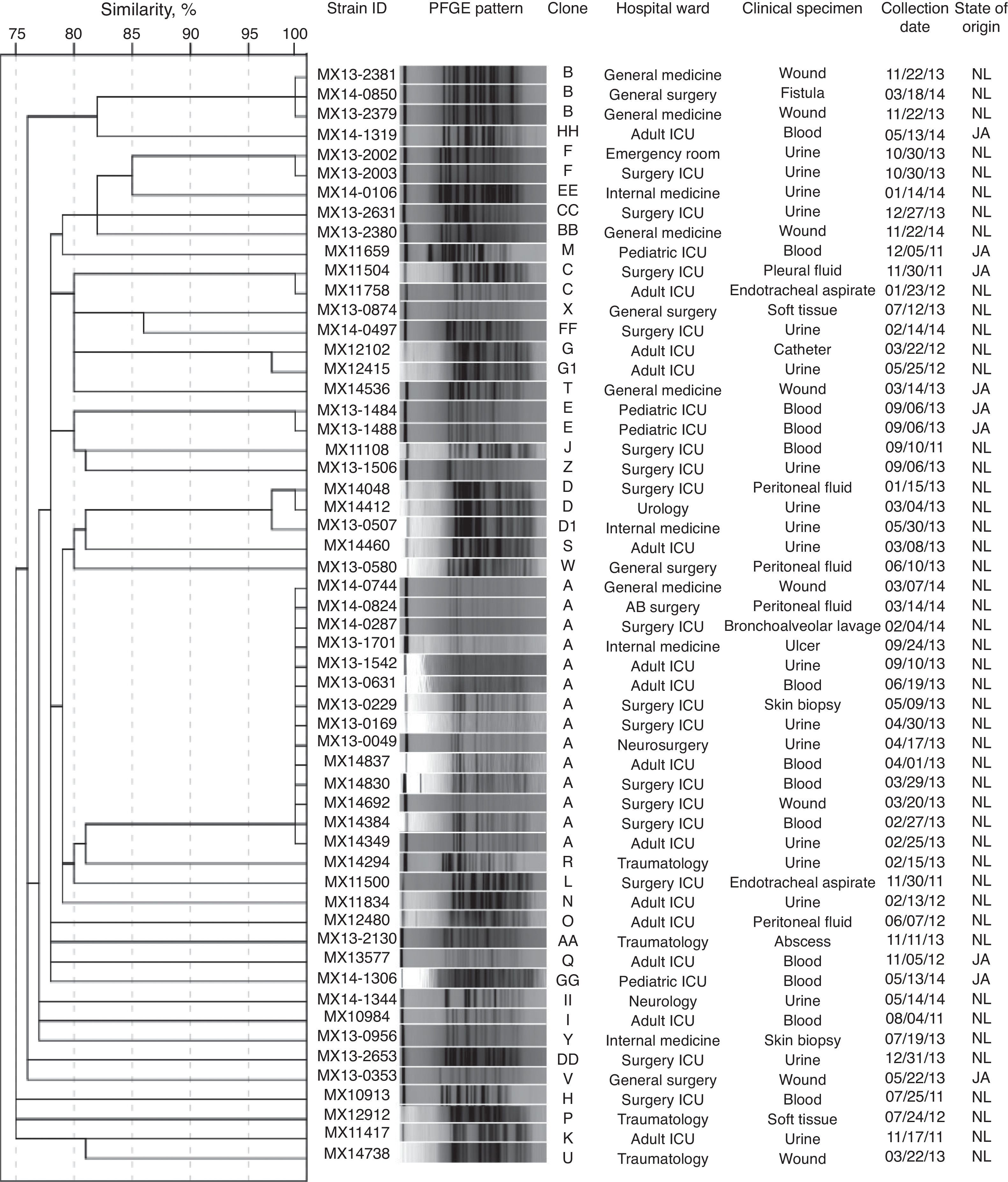
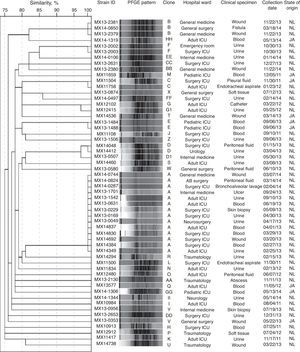
ResultsVREF isolates were highly resistant to vancomycin, erythromycin, norfloxacin, high-level streptomycin, and teicoplanin, and showed lower resistance to tetracycline, nitrofurantoin and quinupristin-dalfopristin. None of the isolates were resistant to linezolid. The vanA gene was detected in all isolates. Two VanB phenotype-vanA genotype isolates, highly resistant to vancomycin and susceptible to teicoplanin, were detected. Furthermore, 17.9% of the isolates were classified as biofilm producers, and the espfm gene was found in 98.2% of the isolates. A total of 37 distinct PFGE patterns and 6 clones (25% of the isolates as clone A, 5.4% as clone B, and 3.6% each as clone C, D, E, and F) were detected. Clone A was detected in 5 different wards of the same hospital during 14 months of surveillance.
ConclusionThe high resistance to most antimicrobial agents and the moderate cross-transmission of VREF detected accentuates the need for continuous surveillance of E. faecium in the hospital setting. This is also the first reported incidence of the E. faecium VanB phenotype-vanA genotype in the Americas.
Enterococcus faecium multifarmacorresistente es un importante patógeno intrahospitalario que a nivel mundial se ha asociado con brotes hospitalarios. El objetivo de este trabajo fue determinar la susceptibilidad a los antimicrobianos, la formación de biopelícula y la relación clonal de los aislamientos clínicos de Enterococcus faecium resistentes a vancomicina (EFRV) en México.
MétodosSe recolectaron 56 aislamientos clínicos en 2 hospitales mexicanos de 2011 a 2014. Los aislamientos de EFRV fueron caracterizados por métodos fenotípicos y moleculares.
ResultadosLos aislamientos de EFRV fueron resistentes a vancomicina, eritromicina, norfloxacina, estreptomicina de alto nivel y teicoplanina. Presentaron baja resistencia a tetraciclina, nitrofurantoína y quinupristina-dalfopristina. Ningún aislamiento presentó resistencia a linezolid. El gen vanA se detectó en todos los aislamientos. Dos aislamientos presentaron un fenotipo VanB-genotipo vanA, que se caracteriza por la resistencia a vancomicina y la susceptibilidad a teicoplanina. El 17,9% de los aislamientos fueron productores de biopelícula y el 98,2% presentaron el gen espfm. Se obtuvieron 37 patrones de bandas diferentes y 6 clonas (25% de la clona A, 5,4% de la clona B y 3,6% de las clonas C, D, E y F, respectivamente). La clona A se detectó en 5 diferentes salas hospitalarias en el mismo hospital durante 14meses.
ConclusiónLa alta resistencia a los antimicrobianos, junto con la moderada transmisión cruzada de EFRV encontradas en este estudio, acentúan, la necesidad de una vigilancia continua de este microorganismo en el ambiente hospitalario. Además, este es el primer reporte de E. faecium con un fenotipo VanB-genotipo vanA en América.
Over the past few decades, Enterococcus species have become one of the most challenging nosocomial problems worldwide due to, in part, the increased use of vancomycin and broad-spectrum antibiotics in hospital. Enterococci can cause a range of nosocomial infections, including urinary tract infections, endocarditis, intra-abdominal and pelvic infections, catheter-related infections, surgical wound infections, and central nervous system infections.1 One of the most common species of Enterococcus involved in nosocomial infections is Enterococcus faecium which is much more frequently resistant to vancomycin and ampicillin than Enterococcus faecalis. Consequently, vancomycin-resistant E. faecium (VREF) represents a growing threat in hospital-acquired infections.2
Enterococci are intrinsically resistant to several antibiotics, and they also readily accumulate mutations and exogenous genes that confer additional resistance. The acquisition of resistance genes often occurs via conjugation with plasmids or conjugative transposons that can potentially carry multiple antibiotic resistance genes.1
Acquired resistance to vancomycin in E. faecium is an increasing problem particularly in hospital-acquired infections. Different genotypes of vancomycin resistance, vanA–G, have been described in enterococci. The vanA and vanB genotypes in E. faecium are most commonly found worldwide, with vanA predominating.3 These genotypes are associated with different levels of antibiotic resistance. The vanA genotype is associated with a high level of vancomycin and teicoplanin resistance, the vanB genotype is associated with a moderate to high level of vancomycin resistance and teicoplanin susceptibility, and the vanC genotype is associated with an intrinsically low level of vancomycin resistance.4
In addition to antibiotic resistance, biofilm formation may be an important factor in the pathogenesis of enterococcal infection. E. faecium is capable of producing biofilms, which consist of a population of cells encased in a hydrated matrix of exopolymeric substances that attach irreversibly to various biotic and abiotic surfaces.5 In fact, enterococci can survive on environmental surfaces, including medical equipment, for long time periods.1 It has been proposed that many environmental and genetic factors are associated with biofilm production, including the enterococcal surface protein, Esp (fm).5,6 Few studies have included an analysis of VREF clinical isolates from Mexican hospitals7,8 and even fewer have reported a molecular analysis of such isolates.7,9–12 Therefore, the aim of this study was to examine the antimicrobial susceptibility, biofilm production, and clonal relatedness of VREF clinical isolates obtained from two tertiary care hospitals in Mexico.
MethodsClinical isolates and study populationFrom 2011 to 2014, consecutive E. faecium isolates were collected at two tertiary care hospitals in Mexico: Hospital Universitario “Dr. José Eleuterio González” in Nuevo Leon and Hospital Civil de Guadalajara and Instituto de Patología Infecciosa y Experimental “Dr. Francisco Ruiz Sánchez” in Jalisco. The Hospital Universitario “Dr. José Eleuterio González” is a 450 bed tertiary-care teaching hospital located in the city of Monterrey, the third largest city in Northeastern Mexico. The hospital provides care to adult and pediatric patients in 20 wards located in a main building and additional services in 3 connected buildings. This hospital serves a population that includes the Monterrey metropolitan area, including 51 municipalities (approximate 5 million Hab.) and surroundings states. During 2013 there were 24,572 discharges, 7667 surgical procedures and a 96.3% daily occupancy rate with patients having a mean length of stay of 4.94 days. The Hospital Civil de Guadalajara Fray Antonio Alcalde is an 899-bed tertiary-care teaching hospital located in the city of Guadalajara, the second largest city in western Mexico. The hospital provides care to adult and pediatric patients in 31 wards situated among 3 connected buildings. This hospital serves a population that includes the greater Guadalajara metropolitan area, including 7 municipalities (approximately 4.0 million Hab) and surroundings states. During 2013 there were 45,982 discharges, 25,415 surgical procedures, and a 94.7% daily occupancy rate with patients having a mean length of stay of 6.2 days.
During the 4-year study period, 151 isolates of E. faecium were collected, and 56 isolates (37.1%) were vancomycin-resistant, of which 48 (85.7%) isolates were from Monterrey and 8 (14.3%) were from Guadalajara. All isolates were considered as presumptive causative agents of infection. One isolate per patient was assayed. The majority of the isolates were recovered from urine (33.9%), followed by blood (23.2%), and soft tissue (14.3%). Most isolates (58.9%) were obtained from intense care unit (ICU) patients. All isolates were cultured on 5% sheep blood agar plates at 37°C for 24h and identified using biochemical tests (i.e., 6.5% NaCl growth, bile esculin hydrolysis, and potassium tellurite reduction). All E. faecium isolates were stored at −80°C until further use. We retrieved demographic data from the patients’ charts as well as information about common risk factors, such as prior antibiotic exposure, hospitalization, and invasive device usage.
Ethics statementThe local ethics committee (Comité de Ética y Comité de Investigación, Facultad de Medicina y Hospital “Dr José Eleuterio González”, Universidad Autónoma de Nuevo León, Mexico) approved this study with the following reference number GA14-015. Informed consent was waived by the Ethics Committee because no intervention was involved and no patient identifying information was included.
Antimicrobial susceptibility and presence of vancomycin resistance-associated genesBroth microdilution susceptibility testing of E. faecium was performed in Sensititre plates (Thermo Scientific-TREK Diagnostic Systems, Ohio, USA) according to the manufacturer's instructions. The antibiotics tested included erythromycin, ampicillin, high-level streptomycin, high-level gentamicin, linezolid, nitrofurantoin, norfloxacin, quinupristin/dalfopristin, tetracycline, teicoplanin, and vancomycin. The results were interpreted according to the Clinical and Laboratory Standards Institute (CLSI) criteria.13 All E. faecium isolates were screened for the presence of vanA, vanB, and vanC as previously described.14
Biofilm production and presence of biofilm production-associated genesBiofilm formation was determined using crystal violet staining as previously described with some modifications.15 All isolates were tested in two different experiments conducted on different days. Cultures diluted 1:40 in 200μl of tryptic soy broth containing 1% glucose were inoculated into 96-well polystyrene plates (Falcon, Franklin Lakes, NJ, USA). After 24h of incubation at 37°C, planktonic cells were removed for determination of cell density at 600nm, and plates were washed 3 times with sterile phosphate buffered saline (PBS), pH 7.3. The adherent biofilms were stained with 0.1% crystal violet for 15min at room temperature, washed 5 times with sterile PBS, and allowed to dry. Finally, an ethanol:acetone (80:20) solution was added to the biofilm samples, and the optical density at 595nm (OD595) was read. The American Type Culture Collection (ATCC) strains, Staphylococcus aureus 29213 (high biofilm producer) and Escherichia coli 25922 (low biofilm producer) were used as controls. Isolates were classified as non-adherent (OD≤0.120), weakly adherent (OD>0.120 to 0.240), or strongly adherent (OD>0.240) according to the criteria described by Christensen et al.16 Determination of the biofilm index was performed using the method previously described by Kristich et al.15 Presence of the espfm gene was determined by PCR using the designed primer, espfm-F (5′-TTGCTAATGCAAGTCCACGTCC-3′) and primer TE105 (5′-GCATCAACACTTGCATTACCGAA-3′), the sequence of which has been previously reported.17 Each 25-μl reaction mixture consisted of 1X PCR buffer, 2mM MgCl2, 0.2mM of each dNTP, 200nM of each primer, 1U of AmpliTaq polymerase (Bioline USA Inc., Randolph, MA, USA) and 2μl of DNA. PCR was initiated by denaturation for 1min at 94°C, followed by 30 cycles of 45s at 94°C, 45s at 58°C and 45s at 72°C with final extension for 1min at 72°C. The PCR products were separated on a 2% agarose gel with a molecular weight standard ladder, and the ethidium bromide-stained bands were visualized by UV transillumination.
Clonal diversity by pulsed-field gel electrophoresis (PFGE)The genetic relatedness of all E. faecium isolates was analyzed by PFGE as previously described with modifications.18 The plugs were incubated overnight in lysis buffer supplemented with RNase (5mg/ml) and lysozyme (1mg/ml), followed by incubation in fresh lysis buffer with proteinase K (0.5mg/ml) at 55°C for 24h. DNA was digested with the restriction enzyme, SmaI, and electrophoresis was performed on the CHEF-DR III system (Bio-Rad Laboratories, Hercules, CA, USA) at 6V/cm, with linear switching interval ramps from 3.5s to 25s for 12h at 14°C for the first block, followed by 1s to 5s for 8h for the second block. Band patterns were visually compared after ethidium bromide staining, and the interpretation was based on the criteria of Tenover et al.19
Statistical analysisStatistical analysis was performed in the SPSS statistics 20.0 software (IBM Corporation, Somers, NY, USA). Demographic and clinical characteristics were compared using Fisher's exact test for percentages and Wilcoxon rank test for continuous variables; a P value of <0.05 was considered as statistically significant. PFGE band patterns were generated by Labworks 4.5 software (Ultra-Violet Products, Upland, CA, USA) with 1% tolerance. The similarity coefficients were generated from a similarity matrix calculated using the Jaccard's coefficient.
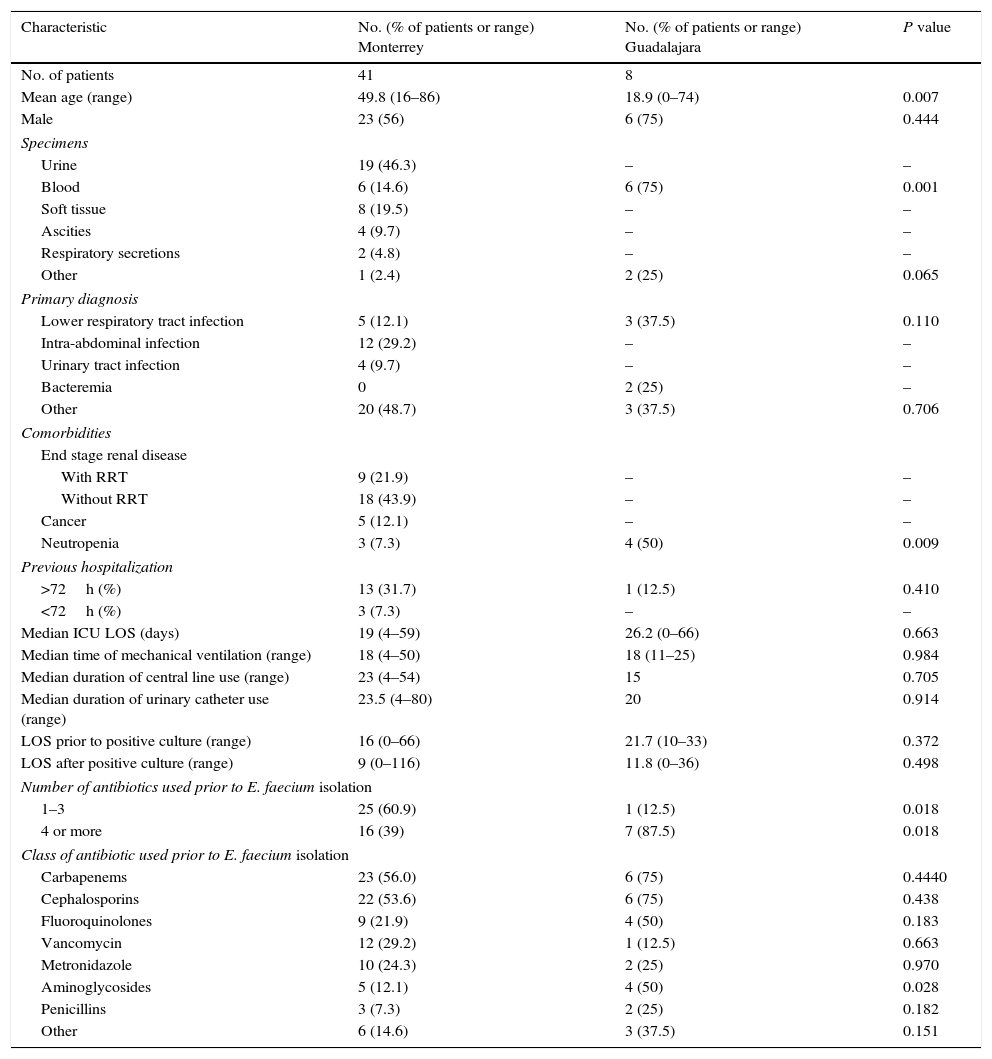
ResultsDemographic and clinical characteristicsWe retrieved demographic data from 49 of 56 patients (41 from Monterrey and 8 from Guadalajara). Patients from Monterrey had a mean age of 49.8 years and were hospitalized mainly for intra-abdominal or lower respiratory tract infections. The majority of patients had been in the ICU with a median stay of 19 days, and 60.9% (n=25) of the patients had received one to three antibiotics before E. faecium was detected. Most patients were treated with carbapenems and cephalosporins. Patients from Guadalajara had a mean age of 44.6 years and were hospitalized mainly for lower respiratory tract or other infections. The majority of patients had been in the ICU with a median stay of 26.2 days, and 87.5% (n=7) of the patients had received at least four antibiotics before E. faecium was detected. Most patients were treated with carbapenems and cephalosporins. There were significant differences in the percentage of isolates from blood, patients with neutropenia and a higher number of patients with prior use of 4 or more antibiotics (especially aminoglycosides) in the group from Guadalajara compared to the group from Monterrey (p<0.05 for all, Table 1).
Demographic and clinical characteristics of patients.
| Characteristic | No. (% of patients or range) Monterrey | No. (% of patients or range) Guadalajara | P value |
|---|---|---|---|
| No. of patients | 41 | 8 | |
| Mean age (range) | 49.8 (16–86) | 18.9 (0–74) | 0.007 |
| Male | 23 (56) | 6 (75) | 0.444 |
| Specimens | |||
| Urine | 19 (46.3) | – | – |
| Blood | 6 (14.6) | 6 (75) | 0.001 |
| Soft tissue | 8 (19.5) | – | – |
| Ascities | 4 (9.7) | – | – |
| Respiratory secretions | 2 (4.8) | – | – |
| Other | 1 (2.4) | 2 (25) | 0.065 |
| Primary diagnosis | |||
| Lower respiratory tract infection | 5 (12.1) | 3 (37.5) | 0.110 |
| Intra-abdominal infection | 12 (29.2) | – | – |
| Urinary tract infection | 4 (9.7) | – | – |
| Bacteremia | 0 | 2 (25) | – |
| Other | 20 (48.7) | 3 (37.5) | 0.706 |
| Comorbidities | |||
| End stage renal disease | |||
| With RRT | 9 (21.9) | – | – |
| Without RRT | 18 (43.9) | – | – |
| Cancer | 5 (12.1) | – | – |
| Neutropenia | 3 (7.3) | 4 (50) | 0.009 |
| Previous hospitalization | |||
| >72h (%) | 13 (31.7) | 1 (12.5) | 0.410 |
| <72h (%) | 3 (7.3) | – | – |
| Median ICU LOS (days) | 19 (4–59) | 26.2 (0–66) | 0.663 |
| Median time of mechanical ventilation (range) | 18 (4–50) | 18 (11–25) | 0.984 |
| Median duration of central line use (range) | 23 (4–54) | 15 | 0.705 |
| Median duration of urinary catheter use (range) | 23.5 (4–80) | 20 | 0.914 |
| LOS prior to positive culture (range) | 16 (0–66) | 21.7 (10–33) | 0.372 |
| LOS after positive culture (range) | 9 (0–116) | 11.8 (0–36) | 0.498 |
| Number of antibiotics used prior to E. faecium isolation | |||
| 1–3 | 25 (60.9) | 1 (12.5) | 0.018 |
| 4 or more | 16 (39) | 7 (87.5) | 0.018 |
| Class of antibiotic used prior to E. faecium isolation | |||
| Carbapenems | 23 (56.0) | 6 (75) | 0.4440 |
| Cephalosporins | 22 (53.6) | 6 (75) | 0.438 |
| Fluoroquinolones | 9 (21.9) | 4 (50) | 0.183 |
| Vancomycin | 12 (29.2) | 1 (12.5) | 0.663 |
| Metronidazole | 10 (24.3) | 2 (25) | 0.970 |
| Aminoglycosides | 5 (12.1) | 4 (50) | 0.028 |
| Penicillins | 3 (7.3) | 2 (25) | 0.182 |
| Other | 6 (14.6) | 3 (37.5) | 0.151 |
RRT=renal replacement therapy; APACHE=acute physiology and chronic health evaluation; LOS=length of stay.
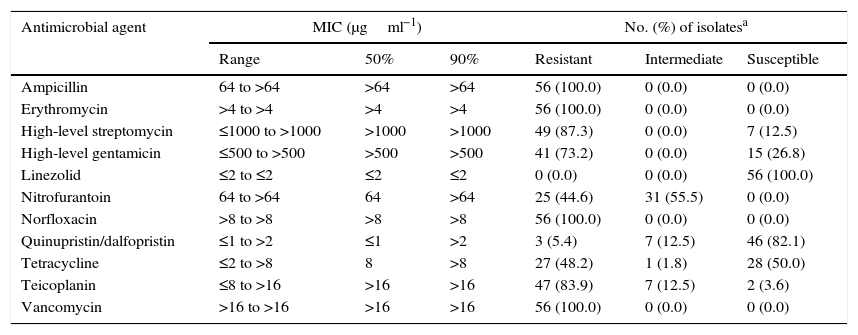
The minimum inhibitory concentration (MIC) range, MIC50, and MIC90, as well as the percentage of isolates resistant, intermediately resistant, and susceptible to each of the antimicrobial agents tested are shown in Table 2. Based on the CLSI interpretive criteria, the E. faecium isolates from Monterrey were highly resistant to vancomycin (100%), ampicillin (100%), erythromycin (100%), norfloxacin (100%), high-level streptomycin (85.4%) and teicoplanin (81.3%). The isolates exhibited lower resistance to high-level gentamicin (68.8%), tetracycline (54.2%), nitrofurantoin (45.8%), and quinupristin/dalfopristin (6.25%). The E. faecium isolates from Guadalajara were highly resistant to vancomycin (100%), teicoplanin (100%), ampicillin (100%), erythromycin (100%), norfloxacin (100%), high-level streptomycin (100%) and high-level gentamicin (100%). The isolates exhibited lower resistance to tetracycline (87.5%), nitrofurantoin (37.5%), and quinupristin/dalfopristin (12.5%). None of the isolates were resistant to linezolid. The vanA gene was detected in all isolates; however, neither vanB nor vanC was detected in any isolate. We also detected two VanB phenotype-vanA genotype isolates (MX13-1542 and MX13-0631), which were highly resistant to vancomycin and susceptible to teicoplanin; these isolates were collected in Monterrey.
Antimicrobial susceptibility of E. faecium isolates.
| Antimicrobial agent | MIC (μgml−1) | No. (%) of isolatesa | ||||
|---|---|---|---|---|---|---|
| Range | 50% | 90% | Resistant | Intermediate | Susceptible | |
| Ampicillin | 64 to >64 | >64 | >64 | 56 (100.0) | 0 (0.0) | 0 (0.0) |
| Erythromycin | >4 to >4 | >4 | >4 | 56 (100.0) | 0 (0.0) | 0 (0.0) |
| High-level streptomycin | ≤1000 to >1000 | >1000 | >1000 | 49 (87.3) | 0 (0.0) | 7 (12.5) |
| High-level gentamicin | ≤500 to >500 | >500 | >500 | 41 (73.2) | 0 (0.0) | 15 (26.8) |
| Linezolid | ≤2 to ≤2 | ≤2 | ≤2 | 0 (0.0) | 0 (0.0) | 56 (100.0) |
| Nitrofurantoin | 64 to >64 | 64 | >64 | 25 (44.6) | 31 (55.5) | 0 (0.0) |
| Norfloxacin | >8 to >8 | >8 | >8 | 56 (100.0) | 0 (0.0) | 0 (0.0) |
| Quinupristin/dalfopristin | ≤1 to >2 | ≤1 | >2 | 3 (5.4) | 7 (12.5) | 46 (82.1) |
| Tetracycline | ≤2 to >8 | 8 | >8 | 27 (48.2) | 1 (1.8) | 28 (50.0) |
| Teicoplanin | ≤8 to >16 | >16 | >16 | 47 (83.9) | 7 (12.5) | 2 (3.6) |
| Vancomycin | >16 to >16 | >16 | >16 | 56 (100.0) | 0 (0.0) | 0 (0.0) |
Ten of the 56 isolates (17.9%) were able to form biofilms. These biofilm-producing isolates were classified as weak biofilm producers (90%) and strong biofilm producers (10%). The espfm gene was found in 98.2% of the isolates. Nine of the isolates (18.8%) obtained in Monterrey were biofilm producers and one of the isolates (12.5%) obtained in Guadalajara was biofilm producer.
Clonal diversityA total of 37 distinct PFGE patterns were identified. The percentage of similarity among the analyzed isolates ranged from 75% to 100%, and the isolates exhibited restriction patterns consisting of 11–20 bands. Isolates that showed 100% similarity in their restriction patterns were classified as a clone. As well, 44.6% (25/56) of the isolates were distributed in 6 clones (A to F), as shown in Fig. 1. Clones A (14/56), B (3/56), C (2/56), D (2/56), and F (2/56) were clusters of isolates obtained from Nuevo Leon. Clone E (2/56) was the only clone obtained from Jalisco. Clone A was detected in 5 different wards of the same hospital during 14 months of surveillance. Among the clone A isolates, 78.6% (11/14) of them were susceptible to high-level gentamicin and resistant to high-level streptomycin (data not shown). We also detected two subtypes (D1 and G1) with restriction patterns that were 97% similar to the D and G patterns, respectively.
DiscussionIn this study, we examined the antimicrobial susceptibility, biofilm production, and clonal relatedness of 56 VREF clinical isolates obtained from two tertiary care hospitals in Mexico during a 4-year study period. Our results demonstrate the presence of VREF isolates that are strongly resistant to vancomycin, erythromycin, norfloxacin, high-level streptomycin, and teicoplanin. Compared to the few previous studies that analyzed VREF clinical isolates from Mexican hospitals7–12 this study included the largest number of VREF clinical isolates to date. Therefore, this work represents a contribution to the national surveillance of emerging nosocomial pathogens.
All the VREF isolates we examined were of the vanA genotype. Similarly, previous studies in Mexico showed that VREF isolates carried the vanA gene,7,9–12 although the vanB gene was also detected.9,11 Interestingly, we detected two VanB phenotype-vanA genotype isolates (MX13-1542 and MX13-0631), which were highly resistant to vancomycin and susceptible to teicoplanin. The VanB phenotype-vanA genotype in E. faecium has been rarely reported worldwide.20–23 In fact, to our knowledge, this is the first detection of this particular E. faecium isolate in the Americas.
As of yet, we do not know the mechanism for the emergence of these VanB phenotype-vanA genotype VREF isolates. The vanA gene cluster is a widely studied vancomycin/teicoplanin resistance determinant which includes 7 van genes (vanA, vanH, vanR, vanS, vanX, vanY, and vanZ) in a Tn1546-type transposon.3,24 It has been reported that the deletion of the intergenic region between vanY and vanZ can partly explain the difference between phenotype and genotype in the VanB phenotype-vanA genotype E. faecium isolate.21 Therefore, further studies should be performed to fully characterize the Tn1546 structure of the two isolates we detected in this study. Tn1546 is generally carried on plasmids and thus is effectively disseminated by horizontal gene transfer. An acquisition of vanA plasmid by a strain of nosocomial E. faecium may result in a spread of VREF infection. Therefore, both characterization of the Tn1546 structure and its linkage to particular plasmid groups is crucial for understanding of VREF dissemination in hospital environments.
There are several risk factors for developing a nosocomial VRE infection, including close physical proximity to patients infected or colonized with VRE, a long period of hospitalization, multiple courses of antimicrobials, hospitalization in intensive-care units and co-morbidities such as diabetes, renal failure or hemodialysis.1 Certainly, the majority of our patients had been in the ICU with a prolonged stay. In addition, half of our patients had received at least four antibiotics before E. faecium was detected, most of them being carbapenems and cephalosporins.
We detected no resistance to linezolid in any of the clinical isolates. Linezolid is one of the two antibiotics that are approved by the Food and Drug Administration (FDA) to treat VREF, and it is used worldwide.1 It has been reported that linezolid resistance in VREF is dependent on prior exposure and duration of therapy.25 Enterococcal infections are commonly treated with ampicillin as the first line treatment, while vancomycin is recommended for cases of ampicillin resistance. Linezolid, daptomycin, and tigecycline are used as alternatives in cases of vancomycin-resistance. Despite linezolid resistance still being rare worldwide1 it has been documented in enterococcal outbreaks26 and even sporadically in patients who have never received the antibiotic.1 Taking these findings and recent reports of linezolid resistant VREF in Mexico11,12 into account, the need for continuous surveillance of E. faecium antimicrobial resistance in hospitals throughout our country is mandatory.
Biofilm formation reportedly occurs less commonly among E. faecium isolates than among E. faecalis isolates, the latter which exhibit a much higher frequency of biofilm formation (95–100%), according to previous reports.27,28 Our result (17.9%) also indicates low biofilm production in E. faecium, and this frequency was even lower than that (53.75%) in a previous report.28 This finding indicates that the clinical impact of these particular VREF isolates is not enhanced by biofilm formation. Furthermore, most of these isolates in our hospitals were obtained from blood and urine specimens, whereas it is thought that biofilm formation is important in conditions such as endocarditis, periodontitis, and a variety of device-related infections5 that were not evident among the patients in our study. Conflicting outcomes regarding the role of the espfm gene in biofilm formation have been published.5 While this gene reportedly plays a role in E. faecalis biofilm formation,29 it is apparently neither essential nor sufficient for biofilm production in infectious E. faecium isolates.27,28 Indeed, we detected a high frequency of the espfm gene in our isolates despite the low level of biofilm production.
Molecular typing methods are essential for identifying hospital-associated outbreaks of E. faecium. Using PFGE, we detected a moderate presence of clones as well as the predominance of clone A, which was detected in 5 different wards of the same hospital during 14 months of surveillance. In addition, the two VanB phenotype-vanA genotype isolates (MX13-1542 and MX13-0631) were detected within the clone A. These two isolates were obtained with a 3-month span from different patients at the ICU. This finding indicates cross-transmission of VREF in the hospital setting. Enterococci can survive for long periods on environmental surfaces, including medical equipment, which may help explain why these organisms were disseminated in the nosocomial setting. In addition, the fact that the two VanB phenotype-vanA genotype isolates were part of clone A underlines the need for continuous monitoring of currently circulating VREF strains to efficiently prevent and control the propagation of nosocomial VREF infections.
One limitation of this study is the lack of using multilocus sequence typing (MLST) for the molecular characterization of E. faecium isolates.30 MLST has revealed the existence of host-specific genogroups, including a specific genetic lineage designated as clonal complex 17 (CC17), which is associated with hospital-related isolates. These strains are characterized by ampicillin and quinolone resistance. The first report of multidrug-resistant VREF isolates in Mexico City that belong to CC17 has been recently published.12 Thus, further studies are needed to analyze our VREF isolates using MLST.
E. faecium isolates resistant to vancomycin, erythromycin, norfloxacin, high-level streptomycin, and teicoplanin exist in Mexican hospitals. The moderate clonal diversity and high frequency of clone A detected among these isolates indicate cross-transmission of VREF in the hospital setting. In addition, this is the first reported incidence of the E. faecium VanB phenotype-vanA genotype in the Americas. This study therefore represents a contribution to the national surveillance of emerging nosocomial pathogens.
Conflict of interestThe authors declare no conflict of interest.
The authors thank the students Estefania Juarez-Tamayaa
Facultad de Química, Universidad Autónoma de Yucatán, Mérida, Yucatán, Mexico.
and Jazmín Dozal-LunabbFacultad de Ciencias Biológicas, Universidad Autónoma de Nuevo León, San Nicolás de los Garza, Nuevo León, Mexico.
for their assistance in the laboratory.