Introduction
Overuse and misuse of antimicrobials has contributed to the emergence of multidrug-resistant (MDR) bacteria, especially in intensive care units. Among MDR bacteria, P. aeruginosa is the second most frequent pathogen associated with pneumonia in hospitalized patients in North America 1 and the most frequent in South America 2. In addition to its relevance as a pathogen, it can acquire resistance to all available antibacterial agents normally used in therapy. Pseudomonas aeruginosa has inherent resistance to many drug classes, can acquire resistance to all relevant treatments via mutations, and can harbour integrons with multiple resistance genes, as those coding for metallo-beta-lactamases, which can cleave the most active antimicrobial agents against P. aeruginosa and Enterobacteriaceae: the carbapenems 3.
The Meropenem Yearly Susceptibility Test Information Collection (MYSTIC) is a global, annual and multi-centre surveillance study that compares the activity of several broad-spectrum antimicrobial agents against bacteria isolated from inpatients at specialized hospital units that are carbapenem users. During the 2002 Brazilian Program's edition, elevated resistance rates in P. aeruginosa were observed to all antimicrobials. As previously described, high resistance level to carbapenems in P. aeruginosa correlates with clonal dissemination 4. This study aimed at investigating intra and inter-centre clonal dissemination of multi-resistant P. aeruginosa responsible for infections in Brazilian intensive care units and to analyse if the high resistance level observed correlates with clonal dissemination.
Methods
Bacterial isolates
Thirty-six multiresistant P. aeruginosa isolates collected during the Brazil MYSTIC Program 2002 from five Intensive Care Units in two Brazilian cities were selected. All the isolates were resistant to meropenem or imipenem plus at least two of the following drugs: ciprofloxacin, cefepime or ceftazidime, piperacillin/tazobactam.
Four centres were located in São Paulo (centres # 1 / 4 / 6 / 7) and one in Brasilia (# 5). Only one isolate per patient was included in the study. The isolates were obtained from different clinical specimens, including urine, respiratory tract secretions, blood specimens and catheter tips.
Identification and susceptibility testing procedures
Bacterial isolates were identified with the GNI VITEK system card (BioMérieux, Inc., Hazelwood, Missouri, USA) and conventional biochemical tests as described elsewhere 5. Susceptibility testing was determined by agar diffusion with Etest (AB BIODISK, Solna, Sweden) according to the manufacturer's procedures. Interpretative criteria used were those described in NCCLS document M100-S14 6.
Phenotypic metallo-beta-lactamase production was investigated by E-test methodology (AB Biodisk, Solna, Sweden), with strips containing imipenem and imipenem plus EDTA. An imipenem/imipenem + EDTA ratio of greater than or equal to eight between the two results was considered as presence of metallo-beta-lactamase. Results were reported as positive (Ratio ≥ 8), negative (Ratio < 8), and non-determinable if dilutions did not allow interpretation as above.
Genotyping
Evaluation of chromosomal polymorphisms was performed by pulsed-field gel electrophoresis (PFGE) as described by Denton et al 7 with minor modifications. Each plug was digested with 10U of SpeI restriction endonuclease (Invitrogen, Carlsbad, CA) at 37 °C for 12 h. Electrophoresis was performed by 1% PFGE agarose gel run on CHEF-DR III system (Bio-Rad Laboratories, Richmond, CA) over 22 h at 14 °C with 5 to 35 s of linear ramping at 6 V/cm. Electrophoretic patterns were analysed with GelCompar II v. 2.5 (Applied Maths, Kortrijik, Belgium) using the interpretative criteria described by Tenover et al 8.
Results
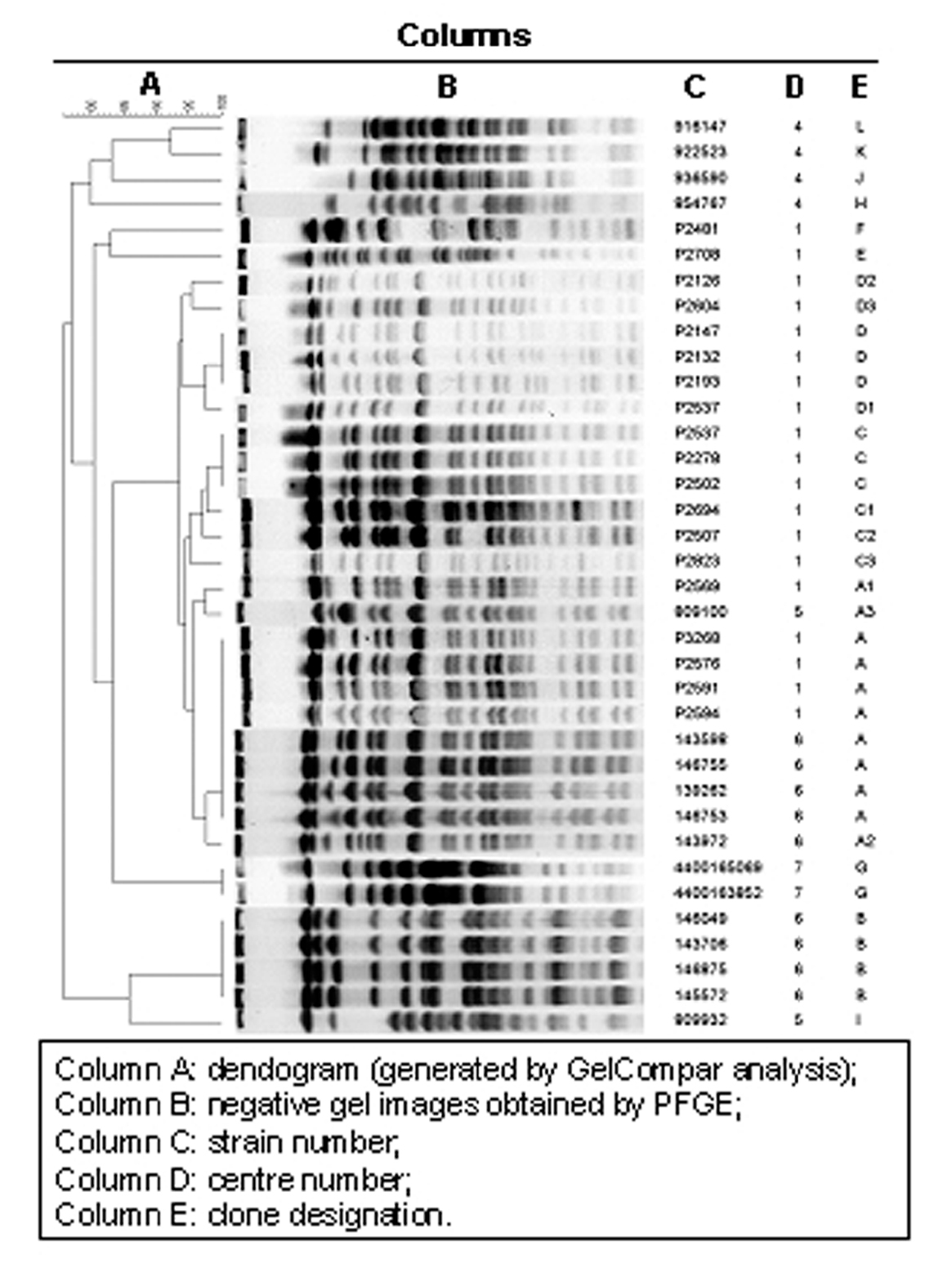
Twenty-two distinct PFGE patterns were observed among 36 isolates. Indistinguishable isolates were interpreted as having the same clonal origin. Five major clones were identified, named A, B, C, D and G (fig. 1). These clones were composed by the following isolates:
Figure 1. Clonal analysis (gel compar analysis).
Eight indistinguishable isolates (14, 25, 26, 41, 42, 48, 62 and 68) belonged to clone A, four isolates (10, 30, 39 and 50) to clone B, three isolates (78, 80 and 101) to clone C, three isolates (6, 8 and 73) to clone D, and two (isolates number 108N and 203N) belonged to clone G.
Furthermore, seven isolates closely related to the above-mentioned clones A, C and D were identified and named A1, A2, A3, C1, C2, C3, and D1. Also, two isolates possibly related to clone D were identified and named respectively D2 and D3. Eight isolates (11, 63, 79, 46AE, 48AE, 53AE, B15 and B19) were unique.
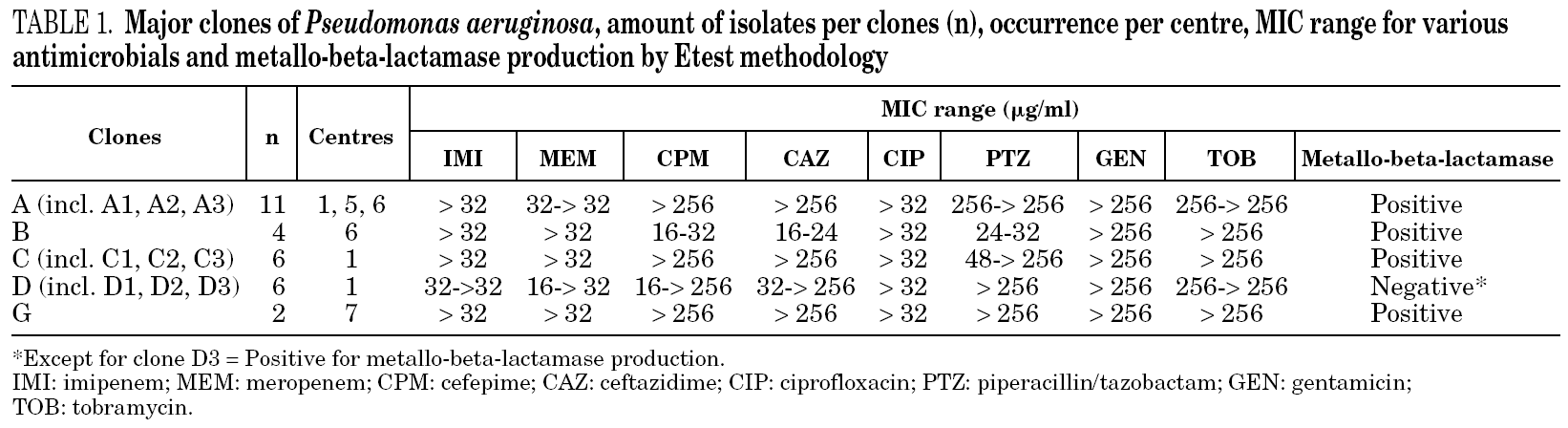
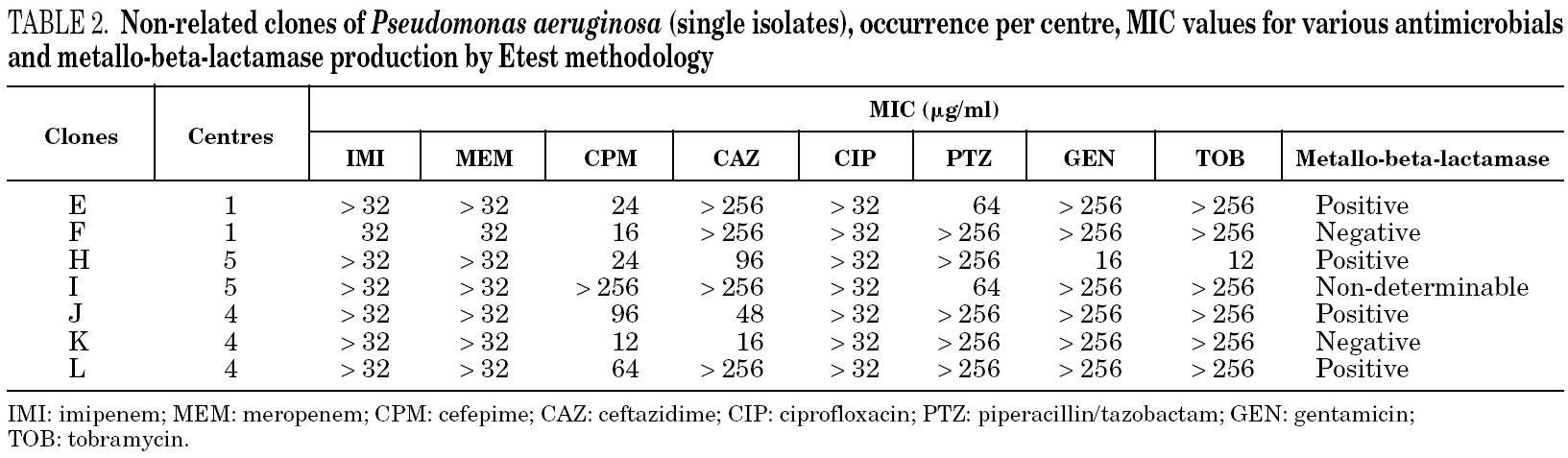
Table 1 shows the minimum inhibitory concentration (MIC) range for imipenem, meropenem, cefepime, ceftazidime, ciprofloxacin, piperacillin/tazobactam, gentamicin, and tobramycin and the results of the metallo-beta-lactamase production test for each of the major clones (i.e. with more than one isolate per group) as determined by Etest methodology, as well as the amount of isolates per clone and their occurrence per centre. Table 2 shows the MIC for the same antimicrobials and the metallo-beta-lactamase production assay for each non-related clone (i.e. with single isolates), as well as their occurrence per centre.
For 28/36 (77.8%) isolates a positive metallo-beta-lactamase production assay was observed. Among the major clones, all but clone D presumably produced metallo-beta-lactamases. Three isolates in clone D, one closely related isolate (clone D1), and one possibly related isolate (clone D2) were not metallo-beta-lactamase producers. However, another possibly related isolate (clone D3) was detected as positive for the production of metallo-beta-lactamase. Among the non-related isolates (i.e. clones with single isolates), 4 tested positive and 2 tested negative for metallo-beta-lactamase production. One isolate (clone I) had a non-determinable result for metallo-beta-lactamase production.
Discussion
Multidrug-resistant P. aeruginosa isolates are usually reported to be responsible for nosocomial infections outbreaks, mainly in intensive care units (ICUs). In this study, 36 multidrug-resistant isolates of P. aeruginosa isolates were clustered in five major genotypes. The presence of a metallo-beta-lactamase was inferred in 28 (77.8%) out of the 36 isolates. No other test, neither phenotypic nor genotypic, to detect the presence of metallo-beta-lactamase was performed in the isolates. Thus, it is only possible to infer the presence of these enzymes, since confirmatory tests would be necessary for precisely determining their production.
MIC results have shown that antimicrobial susceptibility phenotypes did not differ significantly within major clones. Furthermore, metallo-beta-lactamase production results among major clones (A, B, C, D and G) were also consistent. All isolates of clones A, B, C, and G tested positive for metallo-beta-lactamase production. Three isolates of clone D and two of its related clones (D1 and D2) tested negative for the production of metallo-beta-lactamase. However, clone D3 tested positive for metallo-beta-lactamase, suggesting a possible non-chromosomal acquisition of this enzyme. Isolates of one of these genotypes (clone A) were identified in two different hospitals in São Paulo, respectively centres 1 and 6, around five kilometres apart from each other. In three centres (1, 5 and 6) closely related isolates (clones A1, A2 and A3) were also detected. Interestingly, centre 5 is located in Brasilia, situated more than 1,000 kilometres away from centres 1 and 6. However, these institutions are public teaching hospitals and might, in some instances, share patients and professionals along a certain period of time.
Our findings indicate the occurrence of inter-hospital spread of a P. aeruginosa genotype, probably due to transfers of infected patients, share of health care workers, and exchange of medical equipments among institutions. Another possible explanation for the fact might lie on contaminated commercial products commonly used by hospitals, although this is less probable and has not been investigated. The detection of clonal dissemination among different hospitals indicates inadequate nosocomial infection control practices. Those inadequacies might be due to non-adherence to infection control protocols, possibly associated with elevated workload or incorrect disinfection of medical devices. Nevertheless, it should be noted that only non-related MDR P. aeruginosa isolates were recovered from centre 4, a private reference institution located in São Paulo. This finding differs from those of Pellegrino et al 9 who reported clonal dissemination between private and public institutions in Rio de Janeiro.
Many strategies have been proposed to control the emergence of antimicrobial resistance through prescription control: antibiotic cycling, antibiotic formulary restriction, educational programs, prior approval programs, and computer-assisted management 10. These strategies, although useful in many situations, may sometimes be wrongly applied if used solely and as a consequence of surveillance programmes. Prescription control measures are not the methods of choice to uphold emergence of resistance due to clonal dissemination, since the problem usually lies on better performance of infection control practices.
Additionally, the use of certain antimicrobials has been previously linked to the emergence of resistant pathogens, as is the case for ciprofloxacin and MDR P. aeruginosa11. On the other hand, apparently this has not been the case for the carbapenems and MDR P. aeruginosa as shown by Jones et al 4 with the MYSTIC program data. In this situation, the observed high resistance rates to carbapenems were correlated to the persistence of specific clonal spreads. Thus, any analysis for investigating correlation between antimicrobial use and resistance should consider a possible biased result due to clonal dissemination and, preferably, should include a method to identify and exclude related clones from the correlation analysis.
In conclusion, surveillance programme's data may be affected in their susceptibility rates due to clonal dissemination, as was the case of P. aeruginosa collected during the MYSTIC Program Brazil 2002. Molecular epidemiology should complement data generated by these programmes in order to refine data analysis. Ultimately this should maximise the identification of inadequate infection control practices and should improve the prevention of resistance emergence.
Presented in part at the 14th European Congress of Clinical Microbiology and Infectious Diseases, Prague, Czech Republic, 1st to 4th May 2004 [Poster P1107].
MYSTIC Brazil Group: Cássia M. Zocolli, Jorge B. Amarante, Hélio Sader, Suzane Silbert, Jorge D. de Mattos, Marinês D. Martino, Luis F. Camargo, Adília Segura, Julival Ribeiro, Lycia Mimica y Sueli Ykko.